1BGQ
 
 | |
1BXD
 
 | | NMR STRUCTURE OF THE HISTIDINE KINASE DOMAIN OF THE E. COLI OSMOSENSOR ENVZ | | 分子名称: | PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, PROTEIN (OSMOLARITY SENSOR PROTEIN (ENVZ)) | | 著者 | Tanaka, T, Saha, S.K, Tomomori, C, Ishima, R, Liu, D, Tong, K.I, Park, H, Dutta, R, Qin, L, Swindells, M.B, Yamazaki, T, Ono, A.M, Kainosho, M, Inouye, M, Ikura, M. | | 登録日 | 1998-10-02 | | 公開日 | 1999-10-02 | | 最終更新日 | 2023-12-27 | | 実験手法 | SOLUTION NMR | | 主引用文献 | NMR structure of the histidine kinase domain of the E. coli osmosensor EnvZ.
Nature, 396, 1998
|
|
3Q5L
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin | | 分子名称: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-19-{[2-(pyrrolidin-1-yl)ethyl]amino}-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | 著者 | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | 登録日 | 2010-12-28 | | 公開日 | 2011-02-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.65 Å) | | 主引用文献 | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K 213 in the presence of 17-AEP-geldanamycin.
To be Published
|
|
3Q5K
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor | | 分子名称: | 4-[6,6-dimethyl-4-oxo-3-(trifluoromethyl)-4,5,6,7-tetrahydro-1H-indazol-1-yl]-2-{[2-(methylsulfanyl)ethyl]amino}benzamide, Heat shock protein 83-1 | | 著者 | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | 登録日 | 2010-12-28 | | 公開日 | 2011-02-23 | | 最終更新日 | 2024-02-21 | | 実験手法 | X-RAY DIFFRACTION (2.35 Å) | | 主引用文献 | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of an inhibitor
To be Published
|
|
3Q5J
 
 | | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin | | 分子名称: | (4E,6Z,8S,9S,10E,12S,13R,14S,16R)-19-{[3-(dimethylamino)propyl]amino}-13-hydroxy-8,14-dimethoxy-4,10,12,16-tetramethyl-3,20,22-trioxo-2-azabicyclo[16.3.1]docosa-1(21),4,6,10,18-pentaen-9-yl carbamate, Heat shock protein 83-1 | | 著者 | Wernimont, A.K, Tempel, W, Lin, Y.H, Hutchinson, A, MacKenzie, F, Fairlamb, A, Cossar, D, Zhao, Y, Schapira, M, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Ferguson, M.A.J, Hui, R, Pizarro, J.C, Hills, T, Structural Genomics Consortium (SGC) | | 登録日 | 2010-12-28 | | 公開日 | 2011-02-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Crystal structure of the amino-terminal domain of HSP90 from Leishmania major, LMJF33.0312:M1-K213 in the presence of 17-DMAP-geldanamycin.
To be Published
|
|
4URO
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Novobiocin | | 分子名称: | DNA GYRASE SUBUNIT B, NOVOBIOCIN | | 著者 | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | 登録日 | 2014-07-01 | | 公開日 | 2014-07-30 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.59 Å) | | 主引用文献 | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
8SBM
 
 | | Crystal structure of the wild-type Catalytic ATP-binding domain of Mtb DosS | | 分子名称: | 1,2-ETHANEDIOL, GAF domain-containing protein, SODIUM ION, ... | | 著者 | Larson, G, Shi, K, Aihara, H, Bhagi-Damodaran, A. | | 登録日 | 2023-04-03 | | 公開日 | 2023-11-08 | | 最終更新日 | 2023-11-29 | | 実験手法 | X-RAY DIFFRACTION (1.47 Å) | | 主引用文献 | Understanding ATP Binding to DosS Catalytic Domain with a Short ATP-Lid.
Biochemistry, 62, 2023
|
|
4U7O
 
 | | Active histidine kinase bound with ATP | | 分子名称: | AMP PHOSPHORAMIDATE, Histidine protein kinase sensor protein | | 著者 | Cai, Y, Hu, X, Sang, J. | | 登録日 | 2014-07-31 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (2.395 Å) | | 主引用文献 | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
4U7N
 
 | | Inactive structure of histidine kinase | | 分子名称: | Histidine protein kinase sensor protein | | 著者 | Cai, Y, Hu, X, Sang, J. | | 登録日 | 2014-07-31 | | 公開日 | 2015-09-02 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Conformational dynamics of the essential sensor histidine kinase WalK.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
7P2N
 
 | | E.coli GyrB24 with inhibitor LSJ38 (EBL2684) | | 分子名称: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-5-oxidanyl-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | 著者 | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | 登録日 | 2021-07-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Exploring the 5-Substituted 2-Aminobenzothiazole-Based DNA Gyrase B Inhibitors Active against ESKAPE Pathogens.
Acs Omega, 8, 2023
|
|
7P2X
 
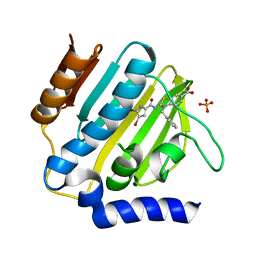 | | E.coli GyrB24 with inhibitor KOB20 (EBL2583) | | 分子名称: | (2Z)-2-[[4,5-bis(bromanyl)-1H-pyrrol-2-yl]carbonylimino]-3-(phenylmethyl)-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | 著者 | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Benek, O, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | 登録日 | 2021-07-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | E.coli GyrB24 with inhibitor KOB20 (EBL2583)
TO BE PUBLISHED
|
|
7P2M
 
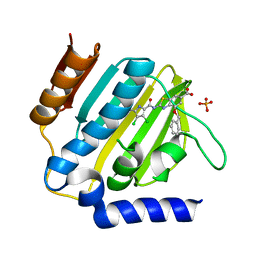 | | E.coli GyrB24 with inhibitor LMD43 (EBL2560) | | 分子名称: | 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, PHOSPHATE ION | | 著者 | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | 登録日 | 2021-07-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.16 Å) | | 主引用文献 | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7P2W
 
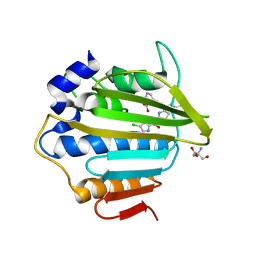 | | E.coli GyrB24 with inhibitor LMD92 (EBL2682) | | 分子名称: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(3-carboxyphenyl)methoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B, ... | | 著者 | Stevenson, C.E.M, Lawson, D.M, Maxwell, A.M, Henderson, S.R, Kikelj, D, Durcik, M, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P. | | 登録日 | 2021-07-06 | | 公開日 | 2022-07-20 | | 最終更新日 | 2024-01-31 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
4URM
 
 | | Crystal Structure of Staph GyraseB 24kDa in complex with Kibdelomycin | | 分子名称: | (1R,4aS,5S,6S,8aR)-5-{[(5S)-1-(3-O-acetyl-4-O-carbamoyl-6-deoxy-2-O-methyl-alpha-L-talopyranosyl)-4-hydroxy-2-oxo-5-(propan-2-yl)-2,5-dihydro-1H-pyrrol-3-yl]carbonyl}-6-methyl-4-methylidene-1,2,3,4,4a,5,6,8a-octahydronaphthalen-1-yl 2,6-dideoxy-3-C-[(1S)-1-{[(3,4-dichloro-5-methyl-1H-pyrrol-2-yl)carbonyl]amino}ethyl]-beta-D-ribo-hexopyranoside, DNA GYRASE SUBUNIT B | | 著者 | Lu, J, Patel, S, Sharma, N, Soisson, S, Kishii, R, Takei, M, Fukuda, Y, Lumb, K.J, Singh, S.B. | | 登録日 | 2014-06-30 | | 公開日 | 2014-07-30 | | 最終更新日 | 2014-10-01 | | 実験手法 | X-RAY DIFFRACTION (2.94 Å) | | 主引用文献 | Structures of Kibdelomycin Bound to Staphylococcus Aureus Gyrb and Pare Showed a Novel U-Shaped Binding Mode.
Acs Chem.Biol., 9, 2014
|
|
1AH8
 
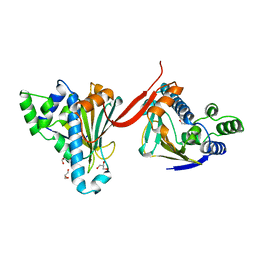 | |
1AH6
 
 | |
1AM1
 
 | |
1A4H
 
 | |
1AJ6
 
 | |
1AMW
 
 | |
1R62
 
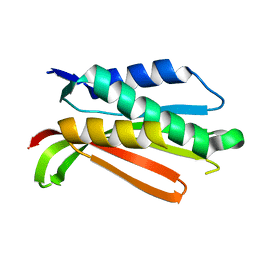 | | Crystal structure of the C-terminal Domain of the Two-Component System Transmitter Protein NRII (NtrB) | | 分子名称: | Nitrogen regulation protein NR(II) | | 著者 | Song, Y, Peisach, D, Pioszak, A.A, Xu, Z, Ninfa, A.J. | | 登録日 | 2003-10-14 | | 公開日 | 2004-06-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Crystal Structure of the C-terminal Domain of the Two-Component System Transmitter Protein Nitrogen Regulator II (NRII; NtrB), Regulator of Nitrogen Assimilation in Escherichia coli.
Biochemistry, 43, 2004
|
|
1S14
 
 | |
2FXS
 
 | | Yeast HSP82 in complex with the novel HSP90 Inhibitor Radamide | | 分子名称: | ATP-dependent molecular chaperone HSP82, GLYCEROL, METHYL 3-CHLORO-2-{3-[(2,5-DIHYDROXY-4-METHOXYPHENYL)AMINO]-3-OXOPROPYL}-4,6-DIHYDROXYBENZOATE | | 著者 | Immormino, R.M, Gewirth, D.T. | | 登録日 | 2006-02-06 | | 公開日 | 2007-02-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Different poses for ligand and chaperone in inhibitor-bound Hsp90 and GRP94: implications for paralog-specific drug design.
J.Mol.Biol., 388, 2009
|
|
7PTF
 
 | | Pseudomonas aeruginosa DNA gyrase B 24kDa ATPase subdomain complexed with novobiocin | | 分子名称: | (4R)-2-METHYLPENTANE-2,4-DIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DNA gyrase subunit B, ... | | 著者 | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | 登録日 | 2021-09-27 | | 公開日 | 2022-10-05 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.32 Å) | | 主引用文献 | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
7PQL
 
 | | Acinetobacter baumannii DNA gyrase B 23kDa ATPase subdomain complexed with EBL2704 | | 分子名称: | 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1R)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, 2-[[3,4-bis(chloranyl)-5-methyl-1H-pyrrol-2-yl]carbonylamino]-4-[(1S)-1-phenylethoxy]-1,3-benzothiazole-6-carboxylic acid, DNA gyrase subunit B | | 著者 | Cotman, A.E, Zega, A, Zidar, N, Ilas, J, Tomasic, T, Masic, L.P, Mundy, J.E.A, Stevenson, C.E.M, Burton, N, Lawson, D.M, Maxwell, A, Kikelj, D. | | 登録日 | 2021-09-17 | | 公開日 | 2022-09-28 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Discovery and Hit-to-Lead Optimization of Benzothiazole Scaffold-Based DNA Gyrase Inhibitors with Potent Activity against Acinetobacter baumannii and Pseudomonas aeruginosa.
J.Med.Chem., 66, 2023
|
|
