1GW6
 
 | | STRUCTURE OF LEUKOTRIENE A4 HYDROLASE D375N MUTANT | | 分子名称: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, ACETATE ION, IMIDAZOLE, ... | | 著者 | Rudberg, P.C, Tholander, F, Thunnissen, M.M.G.M, Samuelsson, B, Haeggstrom, J.Z. | | 登録日 | 2002-03-07 | | 公開日 | 2003-03-06 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Leukotriene A4 Hydrolase: Selective Abrogation of Leukotriene B4 Formation by Mutation of Aspartic Acid 375
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1H19
 
 | | STRUCTURE OF [E271Q]LEUKOTRIENE A4 HYDROLASE | | 分子名称: | ACETIC ACID, IMIDAZOLE, LEUKOTRIENE A-4 HYDROLASE, ... | | 著者 | Rudberg, P.C, Tholander, F, Thunnissen, M.M.G.M, Haeggstrom, J.Z. | | 登録日 | 2002-07-04 | | 公開日 | 2002-08-08 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Leukotriene A4 Hydrolase/Aminopeptidase, Glutamate 271 is a Catalyticresidue with Specific Roles in Two Distinct Enzyme Mechanisms
J.Biol.Chem., 277, 2002
|
|
1HS6
 
 | | STRUCTURE OF LEUKOTRIENE A4 HYDROLASE COMPLEXED WITH BESTATIN. | | 分子名称: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, ACETATE ION, IMIDAZOLE, ... | | 著者 | Thunnissen, M.M.G.M, Nordlund, P.N, Haeggstrom, J.Z. | | 登録日 | 2000-12-24 | | 公開日 | 2001-06-24 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Crystal structure of human leukotriene A(4) hydrolase, a bifunctional enzyme in inflammation.
Nat.Struct.Biol., 8, 2001
|
|
1SQM
 
 | | STRUCTURE OF [R563A] LEUKOTRIENE A4 HYDROLASE | | 分子名称: | ACETIC ACID, IMIDAZOLE, LEUKOTRIENE A-4 HYDROLASE, ... | | 著者 | Tholander, F.O.T, Rudberg, P.C, Thunnissen, M.M.G.M, Haeggstrom, J.Z. | | 登録日 | 2004-03-19 | | 公開日 | 2004-08-03 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Leukotriene A4 hydrolase: identification of a common carboxylate recognition site for the epoxide hydrolase and aminopeptidase substrates
J.Biol.Chem., 279, 2004
|
|
1Z1W
 
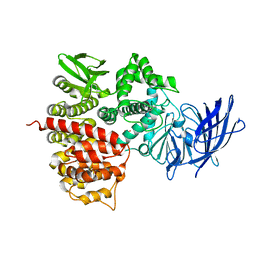 | | Crystal structures of the tricorn interacting facor F3 from Thermoplasma acidophilum, a zinc aminopeptidase in three different conformations | | 分子名称: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | 著者 | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | 登録日 | 2005-03-07 | | 公開日 | 2005-05-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 394, 2005
|
|
1Z5H
 
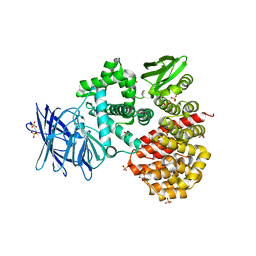 | | Crystal structures of the Tricorn interacting Factor F3 from Thermoplasma acidophilum | | 分子名称: | SULFATE ION, Tricorn protease interacting factor F3, ZINC ION | | 著者 | Kyrieleis, O.J.P, Goettig, P, Kiefersauer, R, Huber, R, Brandstetter, H. | | 登録日 | 2005-03-18 | | 公開日 | 2005-06-28 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Crystal Structures of the Tricorn Interacting Factor F3 from Thermoplasma acidophilum, a Zinc Aminopeptidase in Three Different Conformations
J.MOL.BIOL., 349, 2005
|
|
2DQ6
 
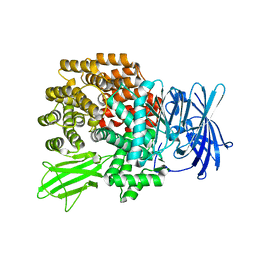 | | Crystal Structure of Aminopeptidase N from Escherichia coli | | 分子名称: | Aminopeptidase N, SULFATE ION, ZINC ION | | 著者 | Nakajima, Y, Onohara, Y, Ito, K, Yoshimoto, T. | | 登録日 | 2006-05-22 | | 公開日 | 2006-08-01 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
2DQM
 
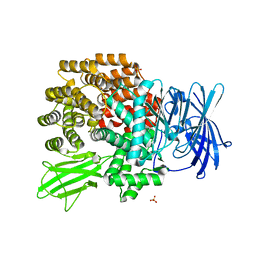 | | Crystal Structure of Aminopeptidase N complexed with bestatin | | 分子名称: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, SULFATE ION, ... | | 著者 | Onohara, Y, Nakajima, Y, Ito, K, Yoshimoto, T. | | 登録日 | 2006-05-29 | | 公開日 | 2006-08-01 | | 最終更新日 | 2023-10-25 | | 実験手法 | X-RAY DIFFRACTION (1.6 Å) | | 主引用文献 | Aminopeptidase N (proteobacteria alanyl aminopeptidase) from Escherichia coli: Crystal structure and conformational change of the methionine 260 residue involved in substrate recognition
J.Biol.Chem., 281, 2006
|
|
2GTQ
 
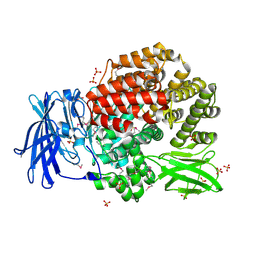 | | Crystal structure of aminopeptidase N from human pathogen Neisseria meningitidis | | 分子名称: | SULFATE ION, ZINC ION, aminopeptidase N | | 著者 | Nocek, B, Mulligan, R, Bargassa, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | 登録日 | 2006-04-28 | | 公開日 | 2006-05-30 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.05 Å) | | 主引用文献 | Crystal structure of aminopeptidase N from human pathogen Neisseria meningitidis.
Proteins, 70, 2007
|
|
2HPO
 
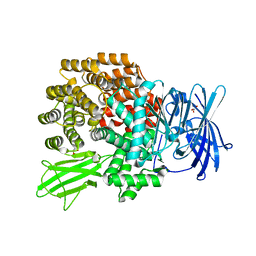 | | Structure of Aminopeptidase N from E. coli Suggests a Compartmentalized, Gated Active Site | | 分子名称: | Aminopeptidase N, GLYCEROL, ZINC ION | | 著者 | Addlagatta, A, Matthews, B.W, Gay, L. | | 登録日 | 2006-07-17 | | 公開日 | 2006-08-15 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.65 Å) | | 主引用文献 | Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2HPT
 
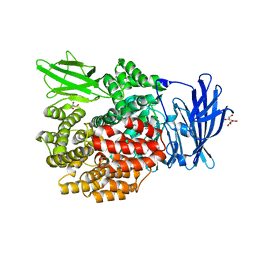 | | Crystal Structure of E. coli PepN (Aminopeptidase N)in complex with Bestatin | | 分子名称: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, Aminopeptidase N, GLYCEROL, ... | | 著者 | Addlagatta, A, Matthews, B.W, Gay, L. | | 登録日 | 2006-07-17 | | 公開日 | 2006-08-15 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Structure of aminopeptidase N from Escherichia coli suggests a compartmentalized, gated active site.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2R59
 
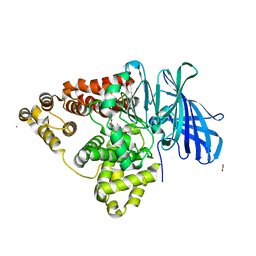 | | Leukotriene A4 hydrolase complexed with inhibitor RB3041 | | 分子名称: | ACETIC ACID, Leukotriene A-4 hydrolase, N-{(2S)-3-[(R)-[(1R)-1-amino-2-phenylethyl](hydroxy)phosphoryl]-2-benzylpropanoyl}-L-phenylalanine, ... | | 著者 | Tholander, F, Haeggstrom, J.Z, Thunnissen, M, Muroya, A, Roques, B.P, Fournie-Zaluski, M.C. | | 登録日 | 2007-09-03 | | 公開日 | 2008-08-12 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.89 Å) | | 主引用文献 | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
2VJ8
 
 | | Complex of human leukotriene A4 hydrolase with a hydroxamic acid inhibitor | | 分子名称: | 6-[{(2S)-2-AMINO-3-[4-(BENZYLOXY)PHENYL]PROPYL}(HYDROXY)AMINO]-6-OXOHEXANOIC ACID), ACETATE ION, IMIDAZOLE, ... | | 著者 | Thunnissen, M.M.G.M, Andersson, B, Wong, C.-H, Samuelsson, B, Haeggstrom, J.Z. | | 登録日 | 2007-12-07 | | 公開日 | 2008-01-15 | | 最終更新日 | 2023-12-13 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Crystal Structures of Leukotriene A4 Hydrolase in Complex with Captopril and Two Competitive Tight-Binding Inhibitors
Faseb J., 16, 2002
|
|
2XPY
 
 | | Structure of Native Leukotriene A4 Hydrolase from Saccharomyces cerevisiae | | 分子名称: | GLUTATHIONE, GLYCEROL, LEUKOTRIENE A-4 HYDROLASE, ... | | 著者 | Helgstrand, C, Hasan, M, Usyal, H, Haeggstrom, J.Z, Thunnissen, M.M.G.M. | | 登録日 | 2010-08-31 | | 公開日 | 2010-12-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.73 Å) | | 主引用文献 | A Leukotriene A(4) Hydrolase-Related Aminopeptidase from Yeast Undergoes Induced Fit Upon Inhibitor Binding.
J.Mol.Biol., 406, 2011
|
|
2XPZ
 
 | | Structure of native yeast LTA4 hydrolase | | 分子名称: | (R,R)-2,3-BUTANEDIOL, LEUKOTRIENE A-4 HYDROLASE, POLYETHYLENE GLYCOL (N=34), ... | | 著者 | Helgstrand, C, Hasan, M, Usyal, H, Haeggstrom, J.Z, Thunnissen, M.M.G.M. | | 登録日 | 2010-08-31 | | 公開日 | 2010-12-29 | | 最終更新日 | 2019-04-24 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | A Leukotriene A(4) Hydrolase-Related Aminopeptidase from Yeast Undergoes Induced Fit Upon Inhibitor Binding.
J.Mol.Biol., 406, 2011
|
|
2XQ0
 
 | | Structure of yeast LTA4 hydrolase in complex with Bestatin | | 分子名称: | 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, CHLORIDE ION, LEUKOTRIENE A-4 HYDROLASE, ... | | 著者 | Helgstrand, C, Hasan, M, Usyal, H, Haeggstrom, J.Z, Thunnissen, M.M.G.M. | | 登録日 | 2010-08-31 | | 公開日 | 2010-12-29 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (1.955 Å) | | 主引用文献 | A Leukotriene A(4) Hydrolase-Related Aminopeptidase from Yeast Undergoes Induced Fit Upon Inhibitor Binding.
J.Mol.Biol., 406, 2011
|
|
2YD0
 
 | | Crystal structure of the soluble domain of human endoplasmic reticulum aminopeptidase 1 ERAP1 | | 分子名称: | 1,2-ETHANEDIOL, 2-(3-AMINO-2-HYDROXY-4-PHENYL-BUTYRYLAMINO)-4-METHYL-PENTANOIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Vollmar, M, Kochan, G, Krojer, T, Ugochukwu, E, Muniz, J.R.C, Raynor, J, Chaikuad, A, Allerston, C, von Delft, F, Bountra, C, Arrowsmith, C.H, Weigelt, J, Edwards, A, Knapp, S. | | 登録日 | 2011-03-17 | | 公開日 | 2011-04-13 | | 最終更新日 | 2020-07-29 | | 実験手法 | X-RAY DIFFRACTION (2.7 Å) | | 主引用文献 | Crystal Structures of the Endoplasmic Reticulum Aminopeptidase-1 (Erap1) Reveal the Molecular Basis for N-Terminal Peptide Trimming.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
2ZXG
 
 | | Aminopeptidase N complexed with the aminophosphinic inhibitor of PL250, a transition state analogue | | 分子名称: | Aminopeptidase N, GLYCEROL, N-{(2S)-3-[(1R)-1-aminoethyl](hydroxy)phosphoryl-2-benzylpropanoyl}-L-phenylalanine, ... | | 著者 | Nakajima, Y, Ito, K, Yoshimoto, T. | | 登録日 | 2008-12-24 | | 公開日 | 2009-08-25 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.55 Å) | | 主引用文献 | Structure of aminopeptidase N from Escherichia coli complexed with the transition-state analogue aminophosphinic inhibitor PL250
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3B2P
 
 | | Crystal structure of E. coli Aminopeptidase N in complex with arginine | | 分子名称: | ARGININE, Aminopeptidase N, GLYCEROL, ... | | 著者 | Anthony, A, Leslie, G, Matthews, B.W. | | 登録日 | 2007-10-18 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (2 Å) | | 主引用文献 | Structural basis for the unusual specificity of Escherichia coli aminopeptidase N.
Biochemistry, 47, 2008
|
|
3B2X
 
 | | Crystal Structure of E. coli Aminopeptidase N in complex with Lysine | | 分子名称: | Aminopeptidase N, GLYCEROL, LYSINE, ... | | 著者 | Addlagatta, A, Gay, L, Matthews, B.W. | | 登録日 | 2007-10-19 | | 公開日 | 2008-05-06 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.5 Å) | | 主引用文献 | Structural basis for the unusual specificity of Escherichia coli aminopeptidase N.
Biochemistry, 47, 2008
|
|
3B34
 
 | |
3B37
 
 | |
3B3B
 
 | |
3B7R
 
 | | Leukotriene A4 Hydrolase Complexed with Inhibitor RB3040 | | 分子名称: | IMIDAZOLE, Leukotriene A-4 hydrolase, N-[3-[(1-AMINOETHYL)(HYDROXY)PHOSPHORYL]-2-(1,1'-BIPHENYL-4-YLMETHYL)PROPANOYL]ALANINE, ... | | 著者 | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | 登録日 | 2007-10-31 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.811 Å) | | 主引用文献 | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
3B7S
 
 | | [E296Q]LTA4H in complex with RSR substrate | | 分子名称: | ACETIC ACID, GLYCEROL, Leukotriene A-4 hydrolase, ... | | 著者 | Tholander, F, Haeggstrom, J, Thunnissen, M, Muroya, A, Roques, B.-P, Fournie-Zaluski, M.-C. | | 登録日 | 2007-10-31 | | 公開日 | 2008-09-16 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.465 Å) | | 主引用文献 | Structure-based dissection of the active site chemistry of leukotriene a4 hydrolase: implications for m1 aminopeptidases and inhibitor design.
Chem.Biol., 15, 2008
|
|
