8ES6
 
 | |
6YHV
 
 | | Structural insights into Pseudomonas aeruginosa Type six secretion system exported effector 8: unliganded Tse8 | | 分子名称: | COPPER (II) ION, Tse8 | | 著者 | Sainz-Polo, M.A, Capuni, R, Pretre, G, Gonzalez-Magana, A, Lucas, M, Altuna, J, Montanchez, I, Fucini, P, Albesa-Jove, D. | | 登録日 | 2020-03-31 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (1.893 Å) | | 主引用文献 | Structural insights into Pseudomonas aeruginosaType six secretion system exported effector 8.
J.Struct.Biol., 212, 2020
|
|
6TE4
 
 | | Structural insights into Pseudomonas aeruginosa Type six secretion system exported effector 8: Tse8 in complex with a peptide | | 分子名称: | Pro-Pro-Leu-Ala-Ser-Lys, Tse8 | | 著者 | Sainz-Polo, M.A, Capuni, R, Lucas, M, Altuna, J, Fucini, P, Montanchez, I, Albesa-Jove, D. | | 登録日 | 2019-11-11 | | 公開日 | 2020-11-04 | | 最終更新日 | 2024-01-24 | | 実験手法 | X-RAY DIFFRACTION (2.29 Å) | | 主引用文献 | Structural insights into Pseudomonas aeruginosaType six secretion system exported effector 8.
J.Struct.Biol., 212, 2020
|
|
6MRG
 
 | | FAAH bound to non covalent inhibitor | | 分子名称: | (1R)-2-{[6-(2,3-dihydro-1,4-benzodioxin-6-yl)pyrimidin-4-yl]amino}-1-phenylethan-1-ol, Fatty-acid amide hydrolase 1 | | 著者 | Saha, A, Shih, A, Mirzadegan, T, Seierstad, M. | | 登録日 | 2018-10-12 | | 公開日 | 2018-10-31 | | 最終更新日 | 2024-03-13 | | 実験手法 | X-RAY DIFFRACTION (2.77 Å) | | 主引用文献 | Predicting the Binding of Fatty Acid Amide Hydrolase Inhibitors by Free Energy Perturbation.
J Chem Theory Comput, 14, 2018
|
|
6KVR
 
 | | Fatty acid amide hydrolase | | 分子名称: | Fatty acid amide hydrolase | | 著者 | Min, C.A, Yun, J.S, Chang, J.H. | | 登録日 | 2019-09-05 | | 公開日 | 2021-09-15 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Comparison of Candida Albicans Fatty Acid Amide Hydrolase Structure with Homologous Amidase Signature Family Enzymes
Crystals, 9, 2019
|
|
6DII
 
 | | Structure of Arabidopsis Fatty Acid Amide Hydrolase in Complex with methyl linolenyl fluorophosphonate | | 分子名称: | Fatty acid amide hydrolase, methyl-9Z,12Z,15Z-octadecatrienylphosphonofluoridate | | 著者 | Aziz, M, Wang, X, Tripathi, A, Bankaitis, V, Chapman, K.D. | | 登録日 | 2018-05-23 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.2 Å) | | 主引用文献 | Structural analysis of a plant fatty acid amide hydrolase provides insights into the evolutionary diversity of bioactive acylethanolamides.
J.Biol.Chem., 294, 2019
|
|
6DHV
 
 | | Structure of Arabidopsis Fatty Acid Amide Hydrolase | | 分子名称: | Fatty acid amide hydrolase | | 著者 | Aziz, M, Wang, X, Tripathi, A, Bankaitis, V, Chapman, K.D. | | 登録日 | 2018-05-21 | | 公開日 | 2019-03-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.099 Å) | | 主引用文献 | Structural analysis of a plant fatty acid amide hydrolase provides insights into the evolutionary diversity of bioactive acylethanolamides.
J.Biol.Chem., 294, 2019
|
|
6C6G
 
 | | An unexpected vestigial protein complex reveals the evolutionary origins of an s-triazine catabolic enzyme. Inhibitor bound complex. | | 分子名称: | AtzG, Biuret hydrolase, CALCIUM ION | | 著者 | Peat, T.S, Esquirol, L, Wilding, M, Liu, J.W, French, N.G, Hartley, C.J, Hideki, O, Easton, C.J, Newman, J, Scott, C. | | 登録日 | 2018-01-18 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | An unexpected vestigial protein complex reveals the evolutionary origins of ans-triazine catabolic enzyme.
J. Biol. Chem., 293, 2018
|
|
6C62
 
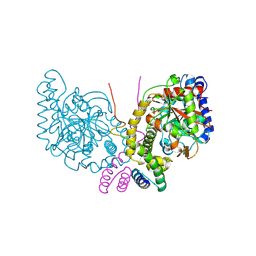 | | An unexpected vestigial protein complex reveals the evolutionary origins of an s-triazine catabolic enzyme. | | 分子名称: | AtzG, Biuret hydrolase, MAGNESIUM ION | | 著者 | Peat, T.S, Esquirol, L, Wilding, M, Liu, J.W, French, N.G, Hartley, C.J, Hideki, O, Easton, C.J, Newman, J, Scott, C. | | 登録日 | 2018-01-17 | | 公開日 | 2018-03-21 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | An unexpected vestigial protein complex reveals the evolutionary origins of ans-triazine catabolic enzyme.
J. Biol. Chem., 293, 2018
|
|
5I8I
 
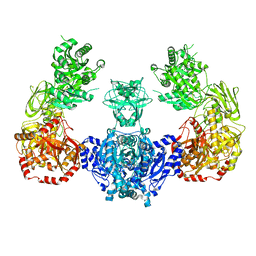 | |
5EWQ
 
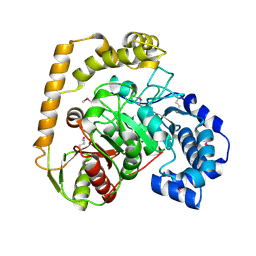 | | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames | | 分子名称: | ACETATE ION, Amidase | | 著者 | Tan, K, Zhou, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | 登録日 | 2015-11-20 | | 公開日 | 2015-12-09 | | 最終更新日 | 2019-12-04 | | 実験手法 | X-RAY DIFFRACTION (2.57 Å) | | 主引用文献 | The crystal structure of an amidase family protein from Bacillus anthracis str. Ames
To Be Published
|
|
5AC3
 
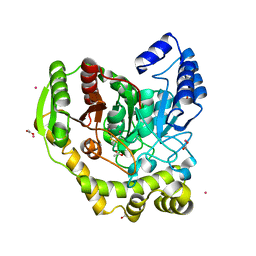 | | Crystal structure of PAM12A | | 分子名称: | ACETIC ACID, CADMIUM ION, PEPTIDE AMIDASE | | 著者 | Wu, B, Wijma, H.J, Song, L, Rozeboom, H.J, Poloni, C, Tian, Y, Arif, M.I, Nuijens, T, Quadflieg, P.J.L.M, Szymanski, W, Feringa, B.L, Janssen, D.B. | | 登録日 | 2015-08-11 | | 公開日 | 2016-07-20 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Versatile Peptide C-Terminal Functionalization Via a Computationally Peptide Amidase
Acs Catalysis, 2016
|
|
4YJI
 
 | | The Crystal Structure of a Bacterial Aryl Acylamidase Belonging to the Amidase signature (AS) enzymes family | | 分子名称: | 3-CYCLOHEXYL-1-PROPYLSULFONIC ACID, Aryl acylamidase, N-(4-HYDROXYPHENYL)ACETAMIDE (TYLENOL) | | 著者 | Choi, I.-G, Lee, S, Park, E.-H, Ko, H.-J, Bang, W.-G. | | 登録日 | 2015-03-03 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-08 | | 実験手法 | X-RAY DIFFRACTION (1.73 Å) | | 主引用文献 | Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family
Biochem.Biophys.Res.Commun., 467, 2015
|
|
4YJ6
 
 | | The Crystal Structure of a Bacterial Aryl Acylamidase Belonging to the Amidase signature (AS) enzymes family | | 分子名称: | Aryl acylamidase, PHOSPHATE ION | | 著者 | Lee, S, Park, E.-H, Ko, H.-J, Bang, W.-G, Choi, I.-G. | | 登録日 | 2015-03-03 | | 公開日 | 2015-11-04 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal structure analysis of a bacterial aryl acylamidase belonging to the amidase signature enzyme family
Biochem.Biophys.Res.Commun., 467, 2015
|
|
4WJ3
 
 | | Crystal structure of the asparagine transamidosome from Pseudomonas aeruginosa | | 分子名称: | 76mer-tRNA, Aspartate--tRNA(Asp/Asn) ligase, Aspartyl/glutamyl-tRNA(Asn/Gln) amidotransferase subunit B, ... | | 著者 | Suzuki, T, Nakamura, A, Kato, K, Tanaka, I, Yao, M. | | 登録日 | 2014-09-29 | | 公開日 | 2014-12-31 | | 最終更新日 | 2020-02-05 | | 実験手法 | X-RAY DIFFRACTION (3.705 Å) | | 主引用文献 | Structure of the Pseudomonas aeruginosa transamidosome reveals unique aspects of bacterial tRNA-dependent asparagine biosynthesis
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4N0I
 
 | | Crystal structure of S. cerevisiae mitochondrial GatFAB in complex with glutamine | | 分子名称: | GLUTAMINE, Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, ... | | 著者 | Araiso, Y, Ishitani, R, Nureki, O. | | 登録日 | 2013-10-02 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.001 Å) | | 主引用文献 | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
4N0H
 
 | | Crystal structure of S. cerevisiae mitochondrial GatFAB | | 分子名称: | Glutamyl-tRNA(Gln) amidotransferase subunit A, mitochondrial, Glutamyl-tRNA(Gln) amidotransferase subunit B, ... | | 著者 | Araiso, Y, Ishitani, R, Nureki, O. | | 登録日 | 2013-10-02 | | 公開日 | 2014-04-16 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (1.952 Å) | | 主引用文献 | Crystal structure of Saccharomyces cerevisiae mitochondrial GatFAB reveals a novel subunit assembly in tRNA-dependent amidotransferases
Nucleic Acids Res., 42, 2014
|
|
4J5P
 
 | | Crystal Structure of a Covalently Bound alpha-Ketoheterocycle Inhibitor (Phenhexyl/Oxadiazole/Pyridine) to a Humanized Variant of Fatty Acid Amide Hydrolase | | 分子名称: | (1S)-1-{5-[5-(bromomethyl)pyridin-2-yl]-1,3-oxazol-2-yl}-7-phenylheptan-1-ol, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | 著者 | Otrubova, K, Brown, M, McCormick, M.S, Han, G.W, O'Neal, S.T, Cravatt, B.F, Stevens, R.C, Lichtman, A.H, Boger, D.L. | | 登録日 | 2013-02-08 | | 公開日 | 2013-05-01 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.3 Å) | | 主引用文献 | Rational design of Fatty Acid amide hydrolase inhibitors that act by covalently bonding to two active site residues.
J.Am.Chem.Soc., 135, 2013
|
|
4IST
 
 | |
4ISS
 
 | |
4HBP
 
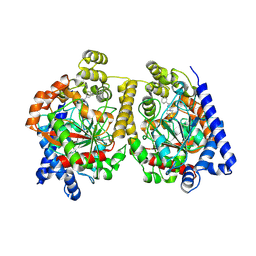 | | Crystal Structure of FAAH in complex with inhibitor | | 分子名称: | 4-(3-phenyl-1,2,4-thiadiazol-5-yl)-N-(pyridin-3-yl)piperazine-1-carboxamide, Fatty-acid amide hydrolase 1 | | 著者 | Behnke, C, Skene, R.J. | | 登録日 | 2012-09-28 | | 公開日 | 2013-02-06 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (2.91 Å) | | 主引用文献 | Synthesis, SAR study, and biological evaluation of a series of piperazine ureas as fatty acid amide hydrolase (FAAH) inhibitors.
Bioorg.Med.Chem., 21, 2013
|
|
4GYS
 
 | |
4GYR
 
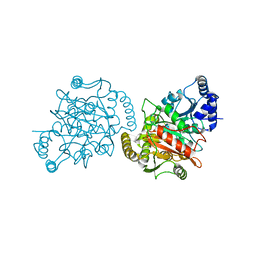 | |
4DO3
 
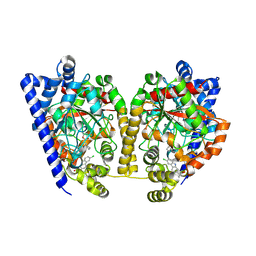 | | Structure of FAAH with a non-steroidal anti-inflammatory drug | | 分子名称: | (2S)-2-(6-chloro-9H-carbazol-2-yl)propanoic acid, CHLORIDE ION, CYCLOHEXANE AMINOCARBOXYLIC ACID, ... | | 著者 | Garau, G. | | 登録日 | 2012-02-09 | | 公開日 | 2013-01-23 | | 最終更新日 | 2023-09-13 | | 実験手法 | X-RAY DIFFRACTION (2.25 Å) | | 主引用文献 | A Binding Site for Nonsteroidal Anti-inflammatory Drugs in Fatty Acid Amide Hydrolase.
J.Am.Chem.Soc., 135, 2013
|
|
4CP8
 
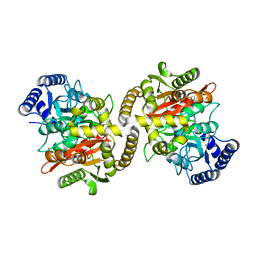 | | Structure of the amidase domain of allophanate hydrolase from Pseudomonas sp strain ADP | | 分子名称: | ALLOPHANATE HYDROLASE, MALONATE ION | | 著者 | Balotra, S, Newman, J, French, N, French, L, Peat, T.S, Scott, C. | | 登録日 | 2014-02-03 | | 公開日 | 2014-11-26 | | 最終更新日 | 2023-12-20 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | X-Ray Structure of the Amidase Domain of Atzf, the Allophanate Hydrolase from the Cyanuric Acid-Mineralizing Multienzyme Complex.
Appl.Environ.Microbiol., 81, 2015
|
|
