4XGU
 
 | | Structure of C. elegans PCH-2 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Putative pachytene checkpoint protein 2, SULFATE ION | | 著者 | Ye, Q, Corbett, K.D. | | 登録日 | 2015-01-02 | | 公開日 | 2015-05-06 | | 最終更新日 | 2024-02-28 | | 実験手法 | X-RAY DIFFRACTION (2.301 Å) | | 主引用文献 | TRIP13 is a protein-remodeling AAA+ ATPase that catalyzes MAD2 conformation switching.
Elife, 4, 2015
|
|
1NSF
 
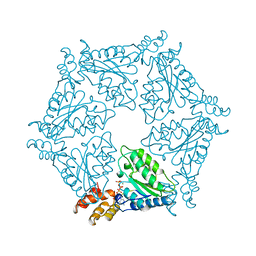 | | D2 HEXAMERIZATION DOMAIN OF N-ETHYLMALEIMIDE SENSITIVE FACTOR (NSF) | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, N-ETHYLMALEIMIDE SENSITIVE FACTOR | | 著者 | Yu, R.C, Hanson, P.I, Jahn, R, Brunger, A.T. | | 登録日 | 1998-06-26 | | 公開日 | 1998-11-25 | | 最終更新日 | 2024-02-14 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP.
Nat.Struct.Biol., 5, 1998
|
|
4YPN
 
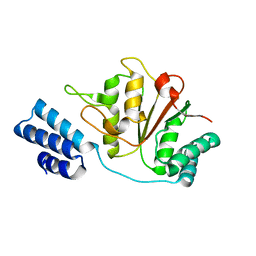 | |
2P65
 
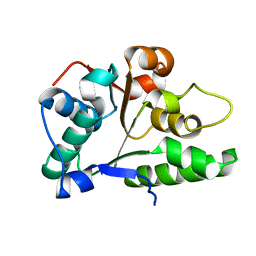 | | Crystal Structure of the first nucleotide binding domain of chaperone ClpB1, putative, (Pv089580) from Plasmodium Vivax | | 分子名称: | Hypothetical protein PF08_0063 | | 著者 | Wernimont, A.K, Lew, J, Kozieradzki, I, Lin, Y.H, Hassanali, A, Zhao, Y, Arrowsmith, C.H, Edwards, A.M, Weigelt, J, Sundstrom, M, Bochkarev, A, Hui, R, Artz, J.D, Structural Genomics Consortium (SGC) | | 登録日 | 2007-03-16 | | 公開日 | 2007-04-03 | | 最終更新日 | 2023-08-30 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Crystal Structure of the first nucleotide binding domain of chaperone ClpB1, putative, (Pv089580) from Plasmodium Vivax
To be Published
|
|
2CHG
 
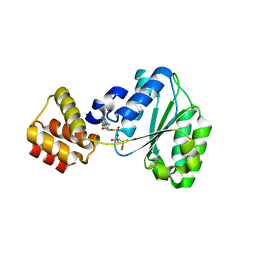 | | Replication Factor C domains 1 and 2 | | 分子名称: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, REPLICATION FACTOR C SMALL SUBUNIT | | 著者 | Seybert, A, Singleton, M.R, Cook, N, Hall, D.R, Wigley, D.B. | | 登録日 | 2006-03-14 | | 公開日 | 2006-06-06 | | 最終更新日 | 2011-07-13 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Communication between Subunits within an Archaeal Clamp-Loader Complex.
Embo J., 25, 2006
|
|
1D2N
 
 | | D2 DOMAIN OF N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN | | 分子名称: | GLYCEROL, MAGNESIUM ION, N-ETHYLMALEIMIDE-SENSITIVE FUSION PROTEIN, ... | | 著者 | Lenzen, C.U, Steinmann, D, Whiteheart, S.W, Weis, W.I. | | 登録日 | 1998-06-30 | | 公開日 | 1998-10-14 | | 最終更新日 | 2024-02-07 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal structure of the hexamerization domain of N-ethylmaleimide-sensitive fusion protein.
Cell(Cambridge,Mass.), 94, 1998
|
|
1JBK
 
 | |
6U1Y
 
 | | bcs1 AAA domain | | 分子名称: | MAGNESIUM ION, Mitochondrial chaperone BCS1, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | 著者 | Tang, W.K, Xia, D. | | 登録日 | 2019-08-17 | | 公開日 | 2020-02-05 | | 最終更新日 | 2024-04-03 | | 実験手法 | X-RAY DIFFRACTION (2.17 Å) | | 主引用文献 | Structures of AAA protein translocase Bcs1 suggest translocation mechanism of a folded protein.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6PB3
 
 | | Structure of Rhizobiales Trip13 | | 分子名称: | Rhizobiales Sp. Pch2, SULFATE ION | | 著者 | Ye, Q, Corbett, K.D. | | 登録日 | 2019-06-12 | | 公開日 | 2019-12-25 | | 最終更新日 | 2020-03-04 | | 実験手法 | X-RAY DIFFRACTION (2.048 Å) | | 主引用文献 | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
7W42
 
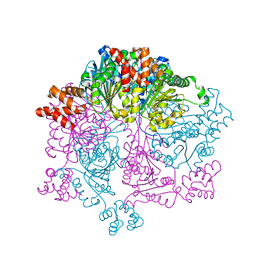 | |
7W46
 
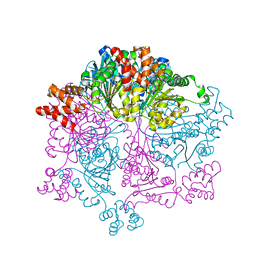 | |
6LK0
 
 | | Crystal structure of human wild type TRIP13 | | 分子名称: | Pachytene checkpoint protein 2 homolog | | 著者 | Wang, Y, Huang, J, Li, B, Xue, H, Tricot, G, Hu, L, Xu, Z, Sun, X, Chang, S, Gao, L, Tao, Y, Xu, H, Xie, Y, Xiao, W, Yu, D, Kong, Y, Chen, G, Sun, X, Lian, F, Zhang, N, Wu, X, Mao, Z, Zhan, F, Zhu, W, Shi, J. | | 登録日 | 2019-12-17 | | 公開日 | 2020-01-22 | | 最終更新日 | 2024-03-27 | | 実験手法 | X-RAY DIFFRACTION (2.6 Å) | | 主引用文献 | A Small-Molecule Inhibitor Targeting TRIP13 Suppresses Multiple Myeloma Progression.
Cancer Res., 80, 2020
|
|
6N2I
 
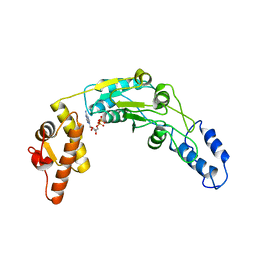 | | Lon protease AAA+ domain | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, DNA-binding ATP-dependent protease La | | 著者 | Botos, I, Li, M, Wlodawer, A, Gustchina, A. | | 登録日 | 2018-11-13 | | 公開日 | 2019-07-10 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (3.5 Å) | | 主引用文献 | New insights into structural and functional relationships between LonA proteases and ClpB chaperones.
Febs Open Bio, 9, 2019
|
|
5VQ9
 
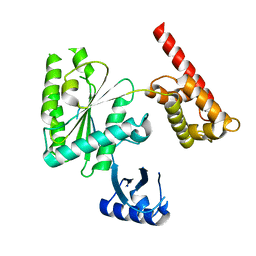 | | Structure of human TRIP13, Apo form | | 分子名称: | Pachytene checkpoint protein 2 homolog | | 著者 | Ye, Q, Corbett, K.D. | | 登録日 | 2017-05-08 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (3.02 Å) | | 主引用文献 | The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding.
EMBO J., 36, 2017
|
|
5VQA
 
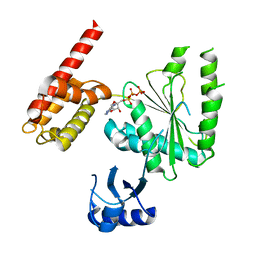 | | Structure of human TRIP13, ATP-bound form | | 分子名称: | ADENOSINE-5'-TRIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | 著者 | Ye, Q, Corbett, K.D. | | 登録日 | 2017-05-08 | | 公開日 | 2017-06-14 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.54 Å) | | 主引用文献 | The AAA+ ATPase TRIP13 remodels HORMA domains through N-terminal engagement and unfolding.
EMBO J., 36, 2017
|
|
5WC2
 
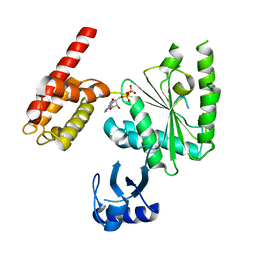 | | Crystal Structure of ADP-bound human TRIP13 | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Pachytene checkpoint protein 2 homolog | | 著者 | Jeong, B.-C, Luo, X. | | 登録日 | 2017-06-29 | | 公開日 | 2018-04-25 | | 最終更新日 | 2023-10-04 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Mechanistic insight into TRIP13-catalyzed Mad2 structural transition and spindle checkpoint silencing.
Nat Commun, 8, 2017
|
|
4W5W
 
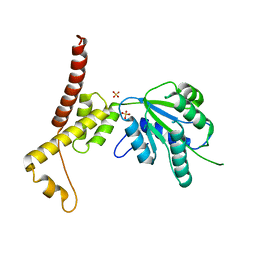 | | Rubisco activase from Arabidopsis thaliana | | 分子名称: | Ribulose bisphosphate carboxylase/oxygenase activase, chloroplastic, SULFATE ION | | 著者 | Hasse, D, Larsson, A.M, Andersson, I. | | 登録日 | 2014-08-19 | | 公開日 | 2015-04-08 | | 最終更新日 | 2024-01-10 | | 実験手法 | X-RAY DIFFRACTION (2.9 Å) | | 主引用文献 | Structure of Arabidopsis thaliana Rubisco activase
Acta Crystallogr.,Sect.D, 71, 2015
|
|
1SXJ
 
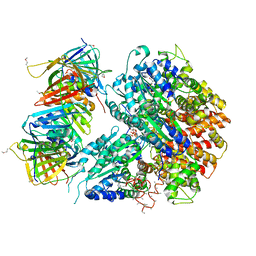 | | Crystal Structure of the Eukaryotic Clamp Loader (Replication Factor C, RFC) Bound to the DNA Sliding Clamp (Proliferating Cell Nuclear Antigen, PCNA) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, Activator 1 37 kDa subunit, Activator 1 40 kDa subunit, ... | | 著者 | Bowman, G.D, O'Donnell, M, Kuriyan, J. | | 登録日 | 2004-03-30 | | 公開日 | 2004-06-22 | | 最終更新日 | 2023-11-15 | | 実験手法 | X-RAY DIFFRACTION (2.85 Å) | | 主引用文献 | Structural analysis of a eukaryotic sliding DNA clamp-clamp loader complex.
Nature, 429, 2004
|
|
5OFO
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state, bound to the model substrate casein | | 分子名称: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | 登録日 | 2017-07-11 | | 公開日 | 2017-08-16 | | 最終更新日 | 2019-11-06 | | 実験手法 | ELECTRON MICROSCOPY (4.6 Å) | | 主引用文献 | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
5OG1
 
 | | Cryo EM structure of the E. coli disaggregase ClpB (BAP form, DWB mutant), in the ATPgammaS state | | 分子名称: | Chaperone protein ClpB,ATP-dependent Clp protease ATP-binding subunit ClpA,Chaperone protein ClpB, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER | | 著者 | Deville, C, Carroni, M, Franke, K.B, Topf, M, Bukau, B, Mogk, A, Saibil, H.R. | | 登録日 | 2017-07-11 | | 公開日 | 2017-08-16 | | 最終更新日 | 2019-08-21 | | 実験手法 | ELECTRON MICROSCOPY (4.5 Å) | | 主引用文献 | Structural pathway of regulated substrate transfer and threading through an Hsp100 disaggregase.
Sci Adv, 3, 2017
|
|
1XWI
 
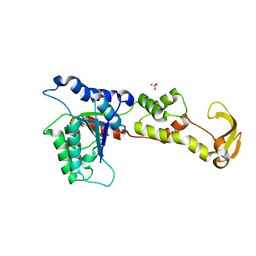 | |
3M6A
 
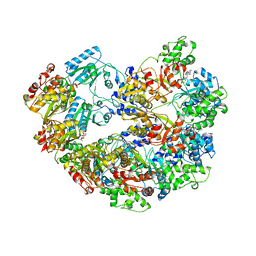 | |
3PXG
 
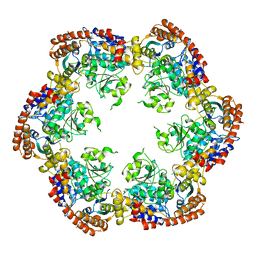 | | Structure of MecA121 and ClpC1-485 complex | | 分子名称: | Adapter protein mecA 1, Negative regulator of genetic competence ClpC/MecB | | 著者 | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | 登録日 | 2010-12-09 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (3.654 Å) | | 主引用文献 | Structure and mechanism of the hexameric MecA-ClpC molecular machine.
Nature, 471, 2011
|
|
3PXI
 
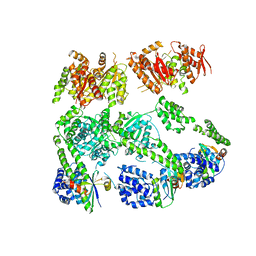 | | Structure of MecA108:ClpC | | 分子名称: | Adapter protein mecA 1, Negative regulator of genetic competence ClpC/MecB | | 著者 | Wang, F, Mei, Z.Q, Wang, J.W, Shi, Y.G. | | 登録日 | 2010-12-09 | | 公開日 | 2011-03-16 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (6.926 Å) | | 主引用文献 | Structure and mechanism of the hexameric MecA-ClpC molecular machine.
Nature, 471, 2011
|
|
4EIW
 
 | | Whole cytosolic region of atp-dependent metalloprotease FtsH (G399L) | | 分子名称: | ADENOSINE-5'-DIPHOSPHATE, ATP-dependent zinc metalloprotease FtsH | | 著者 | Suno, R, Niwa, H, Tsuchiya, D, Yoshida, M, Morikawa, K. | | 登録日 | 2012-04-06 | | 公開日 | 2012-06-06 | | 最終更新日 | 2024-03-20 | | 実験手法 | X-RAY DIFFRACTION (3.9 Å) | | 主引用文献 | Structure of the whole cytosolic region of ATP-dependent protease FtsH
Mol.Cell, 22, 2006
|
|
