7RM8
 
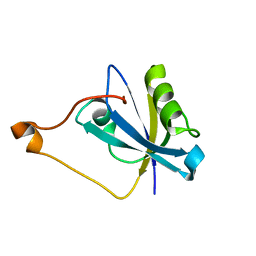 | |
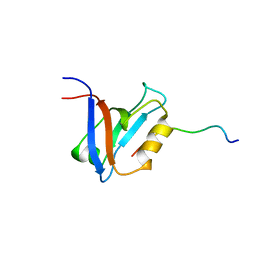
1D5G
 
 | |
8HCK
 
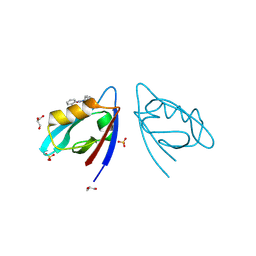 | |
8HU2
 
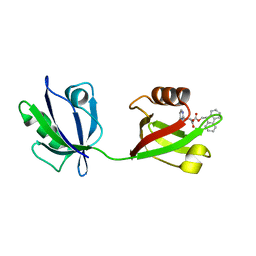 | | Rattus Syntenin-1 PDZ domain with inhibitor | | Descriptor: | (2~{S})-2-(9~{H}-fluoren-9-ylmethoxycarbonylamino)-3-(4-oxidanylidene-5~{H}-pyrimidin-2-yl)propanoic acid, Syntenin-1 | | Authors: | Heo, Y, Lee, J, Yun, J.H, Lee, W. | | Deposit date: | 2022-12-22 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of STNPDZ with inhibitor at 1.60 Angstroms resolution.
To Be Published
|
|
1BFE
 
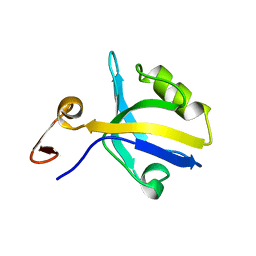 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 | | Descriptor: | PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
1B8Q
 
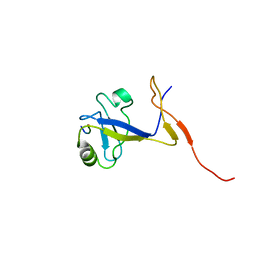 | | SOLUTION STRUCTURE OF THE EXTENDED NEURONAL NITRIC OXIDE SYNTHASE PDZ DOMAIN COMPLEXED WITH AN ASSOCIATED PEPTIDE | | Descriptor: | PROTEIN (HEPTAPEPTIDE), PROTEIN (NEURONAL NITRIC OXIDE SYNTHASE) | | Authors: | Tochio, H, Zhang, Q, Mandal, P, Li, M, Zhang, M. | | Deposit date: | 1999-02-01 | | Release date: | 1999-04-29 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the extended neuronal nitric oxide synthase PDZ domain complexed with an associated peptide.
Nat.Struct.Biol., 6, 1999
|
|
1BE9
 
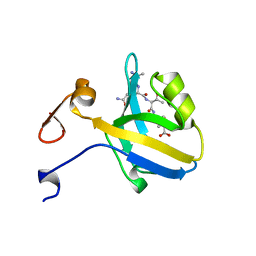 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 IN COMPLEX WITH A C-TERMINAL PEPTIDE DERIVED FROM CRIPT. | | Descriptor: | CRIPT, PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
7CQF
 
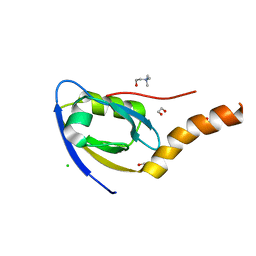 | |
8OEP
 
 | | Crystal structure of the PTPN3 PDZ domain bound to the HPV18 E6 oncoprotein C-terminal peptide | | Descriptor: | Protein E6, SODIUM ION, Tyrosine-protein phosphatase non-receptor type 3 | | Authors: | Genera, M, Colcombet-Cazenave, B, Croitoru, A, Raynal, B, Mechaly, A, Caillet, J, Haouz, A, Wolff, N, Caillet-Saguy, C. | | Deposit date: | 2023-03-11 | | Release date: | 2023-05-10 | | Last modified: | 2023-05-31 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Interactions of the protein tyrosine phosphatase PTPN3 with viral and cellular partners through its PDZ domain: insights into structural determinants and phosphatase activity.
Front Mol Biosci, 10, 2023
|
|
7D6F
 
 | | The crystal structure of ARMS-PBM/MAGI2-PDZ4 | | Descriptor: | GLYCEROL, Kinase D-interacting substrate of 220 kDa, Membrane-associated guanylate kinase, ... | | Authors: | Ye, J, Zhang, Y, Zhong, Z, Wang, C. | | Deposit date: | 2020-09-30 | | Release date: | 2020-11-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.001 Å) | | Cite: | Crystal structure of the PDZ4 domain of MAGI2 in complex with PBM of ARMS reveals a canonical PDZ recognition mode.
Neurochem.Int., 149, 2021
|
|
7E0B
 
 | |
4AMH
 
 | | Influence of circular permutation on the folding pathway of a PDZ domain | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DISKS LARGE HOMOLOG 1, GLYCEROL | | Authors: | Hultqvist, G, Punekar, A.S, Chi, C.N, Selmer, M, Gianni, S, Jemth, P. | | Deposit date: | 2012-03-10 | | Release date: | 2012-12-05 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Tolerance of Protein Folding to a Circular Permutation in a Pdz Domain
Plos One, 7, 2012
|
|
7LUL
 
 | | Structure of the MM2 Erbin PDZ variant in complex with a high-affinity peptide | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Teyra, J, McLaughlin, M, Ernst, A, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2021-02-22 | | Release date: | 2021-07-28 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Comprehensive Assessment of the Relationship Between Site -2 Specificity and Helix alpha 2 in the Erbin PDZ Domain.
J.Mol.Biol., 433, 2021
|
|
7MFW
 
 | |
7NTK
 
 | | PALS1 PDZ1 domain with SARS-CoV-2_E PBM complex | | Descriptor: | 1,2-ETHANEDIOL, CITRATE ANION, Envelope small membrane protein, ... | | Authors: | Javorsky, A, Kvansakul, M. | | Deposit date: | 2021-03-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis of coronavirus E protein interactions with human PALS1 PDZ domain.
Commun Biol, 4, 2021
|
|
7NTJ
 
 | | PALS1 PDZ1 domain with SARS-CoV-1_E PBM complex | | Descriptor: | Envelope small membrane protein, MAGUK p55 subfamily member 5 | | Authors: | Javorsky, A, Kvansakul, M. | | Deposit date: | 2021-03-10 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural basis of coronavirus E protein interactions with human PALS1 PDZ domain.
Commun Biol, 4, 2021
|
|
1G9O
 
 | | FIRST PDZ DOMAIN OF THE HUMAN NA+/H+ EXCHANGER REGULATORY FACTOR | | Descriptor: | NHE-RF | | Authors: | Karthikeyan, S, Leung, T, Birrane, G, Webster, G, Ladias, J.A.A. | | Deposit date: | 2000-11-26 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the PDZ1 domain of human Na(+)/H(+) exchanger regulatory factor provides insights into the mechanism of carboxyl-terminal leucine recognition by class I PDZ domains.
J.Mol.Biol., 308, 2001
|
|
1GM1
 
 | | Second PDZ Domain (PDZ2) of PTP-BL | | Descriptor: | PROTEIN TYROSINE PHOSPHATASE | | Authors: | Walma, T, Tessari, M, Aelen, J, Schepens, J, Hendriks, W, Vuister, G.W. | | Deposit date: | 2001-09-06 | | Release date: | 2002-03-08 | | Last modified: | 2018-03-28 | | Method: | SOLUTION NMR | | Cite: | Structure, dynamics and binding characteristics of the second PDZ domain of PTP-BL.
J. Mol. Biol., 316, 2002
|
|
4E35
 
 | |
4E34
 
 | | Crystal structure of CFTR Associated Ligand (CAL) PDZ domain bound to iCAL36 (ANSRWPTSII) peptide | | Descriptor: | GLYCEROL, Golgi-associated PDZ and coiled-coil motif-containing protein, decameric peptide, ... | | Authors: | Amacher, J.F, Beck, T, Madden, D.R. | | Deposit date: | 2012-03-09 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.395 Å) | | Cite: | Stereochemical Determinants of C-terminal Specificity in PDZ Peptide-binding Domains: A NOVEL CONTRIBUTION OF THE CARBOXYLATE-BINDING LOOP.
J.Biol.Chem., 288, 2013
|
|
4E3B
 
 | |
1GQ4
 
 | |
1GQ5
 
 | |
1I92
 
 | | STRUCTURAL BASIS OF THE NHERF PDZ1-CFTR INTERACTION | | Descriptor: | CHLORIDE ION, NA+/H+ EXCHANGE REGULATORY CO-FACTOR | | Authors: | Karthikeyan, S, Leung, T, Ladias, J.A.A. | | Deposit date: | 2001-03-16 | | Release date: | 2001-06-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis of the Na+/H+ exchanger regulatory factor PDZ1 interaction with the carboxyl-terminal region of the cystic fibrosis transmembrane conductance regulator.
J.Biol.Chem., 276, 2001
|
|
4F8K
 
 | | Molecular analysis of the interaction between the prostacyclin receptor and the first PDZ domain of PDZK1 | | Descriptor: | Na(+)/H(+) exchange regulatory cofactor NHE-RF3, Prostacyclin receptor | | Authors: | Kocher, O, Birrane, G, Kinsella, B.T, Mulvaney, E.P. | | Deposit date: | 2012-05-17 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Molecular Analysis of the Prostacyclin Receptor's Interaction with the PDZ1 Domain of Its Adaptor Protein PDZK1.
Plos One, 8, 2013
|
|
