6CT6
 
 | | Crystal structure of lactate dehydrogenase from Eimeria maxima with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Lactate dehydrogenase, OXAMIC ACID, ... | | Authors: | Wirth, J.D, Xu, C, Theobald, D.L. | | Deposit date: | 2018-03-22 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | The Mechanistic, Structural, and Evolutionary Origin of Lactate Dehydrogenase Substrate Specificity in Apicomplexa
To Be Published
|
|
6TXR
 
 | | Structural insights into cubane-modified aptamer recognition of a malaria biomarker | | Descriptor: | 2'-DEOXYADENOSINE-5'-MONOPHOSPHATE, 2'-DEOXYCYTIDINE-5'-MONOPHOSPHATE, 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, ... | | Authors: | Cheung, Y, Roethlisberger, P, Mechaly, A, Weber, P, Wong, A, Lo, Y, Haouz, A, Savage, P, Hollenstein, M, Tanner, J. | | Deposit date: | 2020-01-14 | | Release date: | 2020-07-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5ES3
 
 | | Co-crystal structure of LDH liganded with oxamate | | Descriptor: | L-lactate dehydrogenase A chain, OXAMIC ACID | | Authors: | Nowicki, M.W, Wear, M.A, McNae, I.W, Blackburn, E.A. | | Deposit date: | 2015-11-16 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | A Streamlined, Automated Protocol for the Production of Milligram Quantities of Untagged Recombinant Rat Lactate Dehydrogenase A Using AKTAxpressTM.
Plos One, 10, 2015
|
|
9LDT
 
 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
9LDB
 
 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
2LDX
 
 | |
6Y91
 
 | | Crystal structure of malate dehydrogenase from Plasmodium Falciparum in complex with NADH | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Romero, A.R, Calderone, V, Gentili, M, Lunev, S, Groves, M, Popowicz, G, Domling, A, Sattler, M. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | A fragment-based approach identifies an allosteric pocket that impacts malate dehydrogenase activity.
Commun Biol, 4, 2021
|
|
6ZZR
 
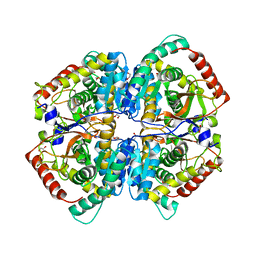 | | The Crystal Structure of human LDHA from Biortus. | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, L-lactate dehydrogenase A chain | | Authors: | Wang, F, Lin, D, Cheng, W, Bao, X, Zhu, B, Shang, H. | | Deposit date: | 2020-08-05 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The Crystal Structure of human LDHA from Biortus
To Be Published
|
|
2PWZ
 
 | |
5NQQ
 
 | |
1B8P
 
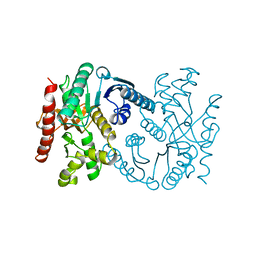 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1BDM
 
 | |
7MDH
 
 | | STRUCTURAL BASIS FOR LIGHT ACITVATION OF A CHLOROPLAST ENZYME. THE STRUCTURE OF SORGHUM NADP-MALATE DEHYDROGENASE IN ITS OXIDIZED FORM | | Descriptor: | PROTEIN (MALATE DEHYDROGENASE), ZINC ION | | Authors: | Johansson, K, Ramaswamy, S, Saarinen, M, Lemaire-Chamley, M, Issakidis-Bourguet, E, Miginiac-Maslow, M, Eklund, H. | | Deposit date: | 1999-02-16 | | Release date: | 1999-06-04 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for light activation of a chloroplast enzyme: the structure of sorghum NADP-malate dehydrogenase in its oxidized form.
Biochemistry, 38, 1999
|
|
5NUF
 
 | | Cytosolic Malate Dehydrogenase 1 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Young, D, Messens, J, Huang, J, Reichheld, J.-P. | | Deposit date: | 2017-04-29 | | Release date: | 2018-01-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis.
J. Exp. Bot., 69, 2018
|
|
5NUE
 
 | | Cytosolic Malate Dehydrogenase 1 (peroxide-treated) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-ETHOXYETHANOL, ... | | Authors: | Young, D, Messens, J, Huang, J, Reichheld, J.-P. | | Deposit date: | 2017-04-29 | | Release date: | 2018-02-28 | | Last modified: | 2018-07-04 | | Method: | X-RAY DIFFRACTION (1.35000277 Å) | | Cite: | Self-protection of cytosolic malate dehydrogenase against oxidative stress in Arabidopsis.
J. Exp. Bot., 69, 2018
|
|
7EPM
 
 | | human LDHC complexed with NAD+ and ethylamino acetic acid | | Descriptor: | 2-(ethylamino)-2-oxidanylidene-ethanoic acid, L-lactate dehydrogenase C chain, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Yu, Y, Chen, Q. | | Deposit date: | 2021-04-27 | | Release date: | 2022-03-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Identification of human LDHC4 as a potential target for anticancer drug discovery.
Acta Pharm Sin B, 12, 2022
|
|
7F8D
 
 | | Malate Dehydrogenase from Geobacillus stearothermophilus (gs-MDH) G218Y mutant | | Descriptor: | Malate dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Shimozawa, Y, Himiyama, T, Nakamura, T, Nishiya, Y. | | Deposit date: | 2021-07-02 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Increasing loop flexibility affords low-temperature adaptation of a moderate thermophilic malate dehydrogenase from Geobacillus stearothermophilus.
Protein Eng.Des.Sel., 34, 2021
|
|
1A5Z
 
 | | LACTATE DEHYDROGENASE FROM THERMOTOGA MARITIMA (TMLDH) | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, CADMIUM ION, L-LACTATE DEHYDROGENASE, ... | | Authors: | Auerbach, G, Ostendorp, R, Prade, L, Korndoerfer, I, Dams, T, Huber, R, Jaenicke, R. | | Deposit date: | 1998-02-18 | | Release date: | 1999-03-23 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lactate dehydrogenase from the hyperthermophilic bacterium thermotoga maritima: the crystal structure at 2.1 A resolution reveals strategies for intrinsic protein stabilization.
Structure, 6, 1998
|
|
1BMD
 
 | |
8J5D
 
 | | Cryo-EM structure of starch degradation complex of BAM1-LSF1-MDH | | Descriptor: | Beta-amylase 1, chloroplastic, Malate dehydrogenase, ... | | Authors: | Guan, Z.Y, Liu, J, Yan, J.J. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The LIKE SEX FOUR 1-malate dehydrogenase complex functions as a scaffold to recruit beta-amylase to promote starch degradation.
Plant Cell, 36, 2023
|
|
7NAY
 
 | | Crystal structure of lactate dehydrogenase from Selenomonas ruminantium with NADH. | | Descriptor: | 1,2-ETHANEDIOL, BORIC ACID, L-lactate dehydrogenase, ... | | Authors: | Bertrand, Q, Robin, A, Girard, E, Madern, D. | | Deposit date: | 2021-01-25 | | Release date: | 2021-02-10 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
1B8V
 
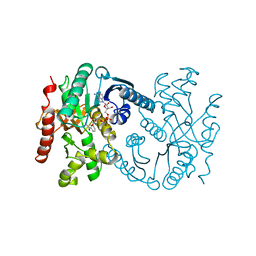 | | Malate dehydrogenase from Aquaspirillum arcticum | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
1B8U
 
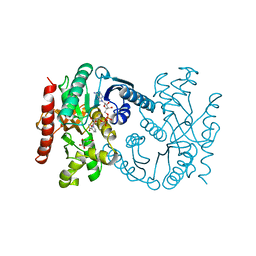 | | MALATE DEHYDROGENASE FROM AQUASPIRILLUM ARCTICUM | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXALOACETATE ION, PROTEIN (MALATE DEHYDROGENASE) | | Authors: | Kim, S.Y, Hwang, K.Y, Kim, S.-H, Han, Y.S, Cho, Y. | | Deposit date: | 1999-02-02 | | Release date: | 1999-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for cold adaptation. Sequence, biochemical properties, and crystal structure of malate dehydrogenase from a psychrophile Aquaspirillium arcticum.
J.Biol.Chem., 274, 1999
|
|
8LDH
 
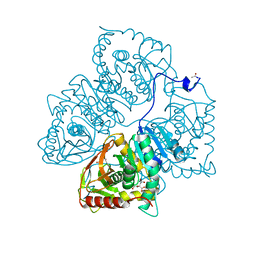 | |
7NRZ
 
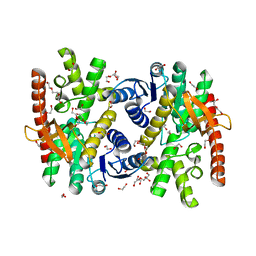 | | Crystal structure of malate dehydrogenase from Trypanosoma cruzi | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Sonani, R.R, Kurpiewska, K, Lewinski, K, Dubin, G. | | Deposit date: | 2021-03-04 | | Release date: | 2022-02-16 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Distinct sequence and structural feature of trypanosoma malate dehydrogenase.
Biochem.Biophys.Res.Commun., 557, 2021
|
|
