8TFF
 
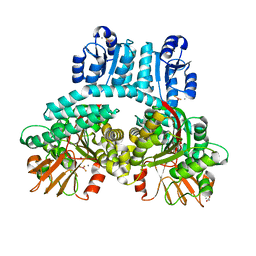 | | PorX with BeF3 phosphate analog | | Descriptor: | ACETATE ION, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Saran, A, Zeytuni, N. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | PorX with BeF3 phosphate analog
To Be Published
|
|
8TFH
 
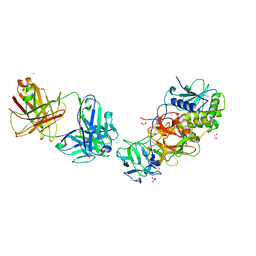 | | Ricin in complex with Fab JB4 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, JB4 monoclonal antibody heavy chain, ... | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Structural Basis of Antibody-Mediated Inhibition of Ricin Toxin Attachment to Host Cells.
Biochemistry, 62, 2023
|
|
8TFL
 
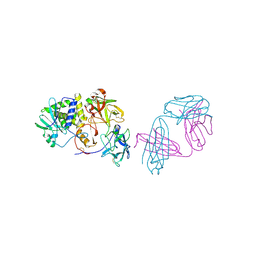 | | Ricin in complex with Fab SylH3 | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, NONAETHYLENE GLYCOL, ... | | Authors: | Rudolph, M.J, Mantis, N. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structural Basis of Antibody-Mediated Inhibition of Ricin Toxin Attachment to Host Cells.
Biochemistry, 62, 2023
|
|
8TFM
 
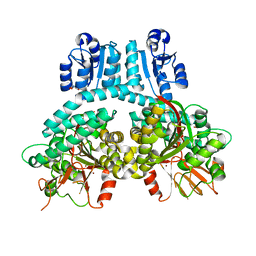 | |
8TGO
 
 | | Crystal structure of the BG505 triple tandem trimer gp140 HIV-1 Env in complex with PGT124 and 35O22 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 35O22 scFv, ... | | Authors: | Xian, Y, Yuan, M, Wilson, I.A. | | Deposit date: | 2023-07-12 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (5.75 Å) | | Cite: | Triple tandem trimer immunogens for HIV-1 and influenza nucleic acid-based vaccines.
Npj Vaccines, 9, 2024
|
|
8THP
 
 | |
8TLZ
 
 | |
8TM0
 
 | |
8TM2
 
 | |
8TO2
 
 | | Bottom cylinder of high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Allophycocyanin subunit alpha-B, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8TO5
 
 | | Central rod disk in C1 symmetry of high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-02 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (1.87 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8TPJ
 
 | | Top cylinder bound to OCP from high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | Allophycocyanin alpha chain, Allophycocyanin beta chain, Orange carotenoid-binding protein, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-04 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.1 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8TRO
 
 | | Rod from high-resolution phycobilisome quenched by OCP (local refinement) | | Descriptor: | C-phycocyanin alpha subunit, C-phycocyanin beta subunit, PHYCOCYANOBILIN, ... | | Authors: | Sauer, P.V, Sutter, M, Cupellini, L. | | Deposit date: | 2023-08-09 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | Structural and quantum chemical basis for OCP-mediated quenching of phycobilisomes.
Sci Adv, 10, 2024
|
|
8TSH
 
 | |
8TSI
 
 | |
8TSL
 
 | |
8TSW
 
 | | S. thermodepolymerans KpsMT-KpsE Apo 1 | | Descriptor: | ABC transporter ATP-binding protein, Capsular biosynthesis protein, Transport permease protein | | Authors: | Kuklewicz, J, Zimmer, J. | | Deposit date: | 2023-08-12 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular insights into capsular polysaccharide secretion.
Nature, 2024
|
|
8TT3
 
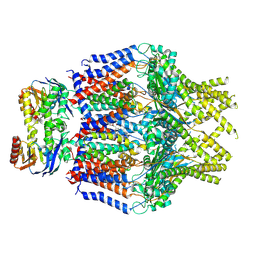 | | S. thermodepolymerans KpsM-KpsE in Glycolipid 2 state with rigid body fitted KpsT | | Descriptor: | (2R,5S,8S)-2,5-dihydroxy-5,10-dioxo-8-[(undecanoyloxy)methyl]-4,6,9-trioxa-5lambda~5~-phosphahenicosan-1-yl 3-deoxy-alpha-L-altro-oct-2-ulopyranosidonic acid, ABC transporter ATP-binding protein, Capsular biosynthesis protein, ... | | Authors: | Kuklewicz, J, Zimmer, J. | | Deposit date: | 2023-08-12 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular insights into capsular polysaccharide secretion.
Nature, 2024
|
|
8TUN
 
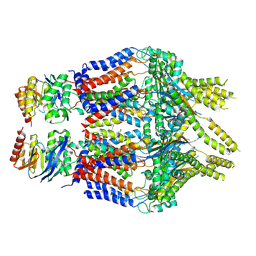 | | S. thermodepolymerans KpsM-KpsE in Glycolipid 1 state with rigid body fitted KpsT | | Descriptor: | (2R,5S,8S)-2,5-dihydroxy-5,10-dioxo-8-[(undecanoyloxy)methyl]-4,6,9-trioxa-5lambda~5~-phosphahenicosan-1-yl 3-deoxy-alpha-L-altro-oct-2-ulopyranosidonic acid, ABC transporter ATP-binding protein, Capsular biosynthesis protein, ... | | Authors: | Kuklewicz, J, Zimmer, J. | | Deposit date: | 2023-08-16 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Molecular insights into capsular polysaccharide secretion.
Nature, 2024
|
|
8TV6
 
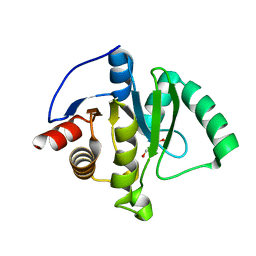 | | SARS-CoV-2 Mac1 in complex with MDOLL-0169 | | Descriptor: | (1R,6R)-6-{[3-(methoxycarbonyl)-5,6,7,8-tetrahydro-4H-cyclohepta[b]thiophen-2-yl]carbamoyl}cyclohex-3-ene-1-carboxylic acid, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Wazir, S, Maksimainen, M, Lehtio, L. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of 2-amide-3-methylester thiophenes inhibiting SARS-CoV-2 ADP-ribosyl hydrolysing macrodomain and coronavirus replication
To Be Published
|
|
8TWQ
 
 | |
8U6Y
 
 | | Preholo-Proteasome from Beta 3 D205 deletion | | Descriptor: | Proteasome assembly chaperone 2, Proteasome chaperone 1, Proteasome maturation factor UMP1, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Velez, B, Blickling, M, Razi, A, Hanna, J. | | Deposit date: | 2023-09-14 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of autocatalytic activation during proteasome assembly
Nat.Struct.Mol.Biol., 2024
|
|
8U7U
 
 | | Proteasome 20S Core Particle from Beta 3 D205 deletion | | Descriptor: | Proteasome subunit alpha type-1, Proteasome subunit alpha type-2, Proteasome subunit alpha type-3, ... | | Authors: | Walsh Jr, R.M, Rawson, S, Velez, B, Blickling, M, Razi, A, Hanna, J. | | Deposit date: | 2023-09-15 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (2.16 Å) | | Cite: | Mechanism of autocatalytic activation during proteasome assembly
Nat.Struct.Mol.Biol., 2024
|
|
8UDC
 
 | | Crystal structure of TcPINK1 in complex with CYC116 | | Descriptor: | (4P)-4-(2-amino-4-methyl-1,3-thiazol-5-yl)-N-[4-(morpholin-4-yl)phenyl]pyrimidin-2-amine, DI(HYDROXYETHYL)ETHER, SULFATE ION, ... | | Authors: | Veyron, S, Rasool, S, Trempe, J.F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural Characterization of a small-molecule inhibitor of PINK1, a precursor tool compound for the study of Parkinson's disease
To Be Published
|
|
8USW
 
 | | CNQX-bound GluN1a-3A NMDA receptor | | Descriptor: | 7-nitro-2,3-dioxo-1,2,3,4-tetrahydroquinoxaline-6-carbonitrile, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Michalski, K, Furukawa, H. | | Deposit date: | 2023-10-30 | | Release date: | 2024-04-17 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Structure and function of GluN1-3A NMDA receptor excitatory glycine receptor channel
To Be Published
|
|
