4XHA
 
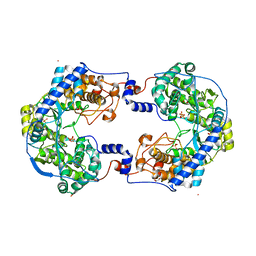 | | Crystal structure of Thosea asigna virus RNA-dependent RNA polymerase (RdRP) complexed with Lu3+ | | Descriptor: | GLYCEROL, LUTETIUM (III) ION, RNA-dependent RNA polymerase, ... | | Authors: | Ferrero, D.S, Buxaderas, M, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2015-01-05 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Structure of the RNA-Dependent RNA Polymerase of a Permutotetravirus Suggests a Link between Primer-Dependent and Primer-Independent Polymerases.
Plos Pathog., 11, 2015
|
|
4XHB
 
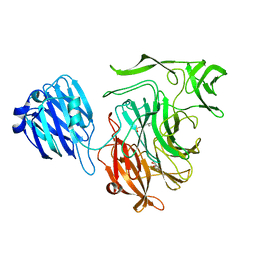 | |
4XHC
 
 | | rhamnosidase from Klebsiella oxytoca with rhamnose bound | | Descriptor: | Alpha-L-rhamnosidase, SULFATE ION, alpha-L-rhamnopyranose | | Authors: | O'Neill, E.O, Stevenson, C.E.M, Patterson, M.J, Rejzek, M, Chauvin, A, Lawson, D.M, Field, R.A. | | Deposit date: | 2015-01-05 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of a novel two domain GH78 family alpha-rhamnosidase from Klebsiella oxytoca with rhamnose bound.
Proteins, 83, 2015
|
|
4XHD
 
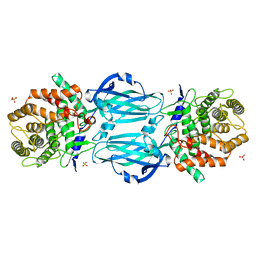
 | | STRUCTURE OF HUMAN PREGNANE X RECEPTOR LIGAND BINDING DOMAIN WITH COMPOUND-1 | | Descriptor: | GLYCEROL, N-{(2R)-1-[(4S)-4-(4-chlorophenyl)-4-hydroxy-3,3-dimethylpiperidin-1-yl]-3-methyl-1-oxobutan-2-yl}-2-cyclopropylacetamide, Nuclear receptor subfamily 1 group I member 2 | | Authors: | Khan, J.A, Camac, D.M. | | Deposit date: | 2015-01-05 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Developing Adnectins That Target SRC Co-Activator Binding to PXR: A Structural Approach toward Understanding Promiscuity of PXR.
J.Mol.Biol., 427, 2015
|
|
4XHE
 
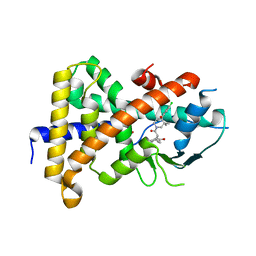
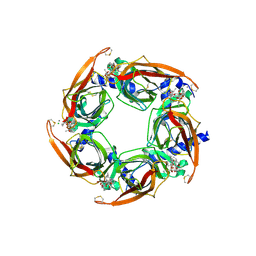 | | Crystal Structure of A-AChBP in complex with pinnatoxin A | | Descriptor: | CALCIUM ION, CHLORIDE ION, Pinnatoxin A, ... | | Authors: | Bourne, Y, Sulzenbacher, G, Marchot, P. | | Deposit date: | 2015-01-05 | | Release date: | 2015-06-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Marine Macrocyclic Imines, Pinnatoxins A and G: Structural Determinants and Functional Properties to Distinguish Neuronal alpha 7 from Muscle alpha 12 beta gamma delta nAChRs.
Structure, 23, 2015
|
|
4XHF
 
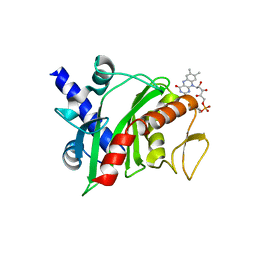 | | Crystal structure of Shewanella oneidensis NqrC | | Descriptor: | FLAVIN MONONUCLEOTIDE, Na-translocating NADH-quinone reductase subunit C NqrC, SODIUM ION | | Authors: | Tomchick, D.R, Brautigam, C.A, Deka, R.K, Norgard, M.V. | | Deposit date: | 2015-01-05 | | Release date: | 2015-12-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Molecular insights into the enzymatic diversity of flavin-trafficking protein (Ftp; formerly ApbE) in flavoprotein biogenesis in the bacterial periplasm.
Microbiologyopen, 5, 2016
|
|
4XHG
 
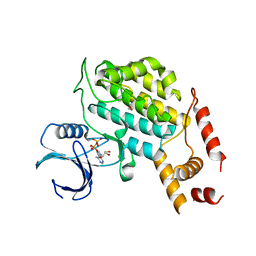 | | Structure of C. glabrata Hrr25 bound to ADP (formate condition) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, FORMIC ACID, Similar to uniprot|P29295 Saccharomyces cerevisiae YPL204w HRR25 | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-01-05 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
4XHH
 
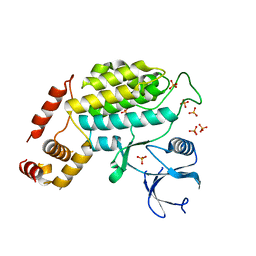 | | Structure of C. glabrata Hrr25, Apo state | | Descriptor: | PHOSPHATE ION, Similar to uniprot|P29295 Saccharomyces cerevisiae YPL204w HRR25 | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-01-05 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
4XHI
 
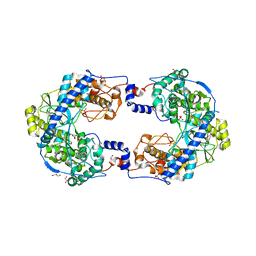 | | Crystal structure of native Thosea asigna virus RNA-dependent RNA polymerase (RdRP) at 2.15 Angstrom resolution | | Descriptor: | GLYCEROL, RNA-dependent RNA polymerase, SULFATE ION | | Authors: | Ferrero, D.S, Buxaderas, M, Rodriguez, J.F, Verdaguer, N. | | Deposit date: | 2015-01-05 | | Release date: | 2015-11-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Structure of the RNA-Dependent RNA Polymerase of a Permutotetravirus Suggests a Link between Primer-Dependent and Primer-Independent Polymerases.
Plos Pathog., 11, 2015
|
|
4XHJ
 
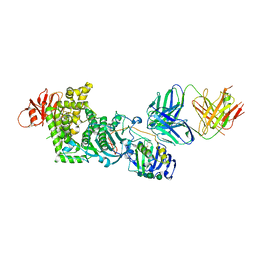 | | gHgL of Varicella-zoster virus in complex with human neutralizing antibodies. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein H, ... | | Authors: | Xing, Y. | | Deposit date: | 2015-01-05 | | Release date: | 2015-05-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.156 Å) | | Cite: | A site of varicella-zoster virus vulnerability identified by structural studies of neutralizing antibodies bound to the glycoprotein complex gHgL.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4XHK
 
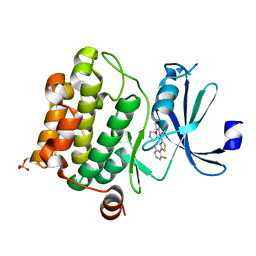 | | PIM1 kinase in complex with Compound 1s | | Descriptor: | 2-(2,6-difluorophenyl)-N-{4-[(3S)-pyrrolidin-3-yloxy]pyridin-3-yl}-1,3-thiazole-4-carboxamide, Serine/threonine-protein kinase pim-1 | | Authors: | Marcotte, D.J, Silvian, L.F. | | Deposit date: | 2015-01-05 | | Release date: | 2015-02-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based design of low-nanomolar PIM kinase inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
4XHL
 
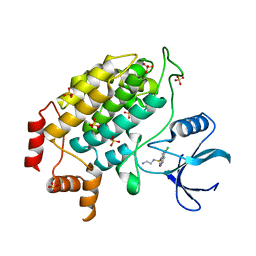 | | Structure of S. cerevisiae Hrr25 1-394 (K38R mutant) | | Descriptor: | Casein kinase I homolog HRR25, N-(2-AMINOETHYL)-5-CHLOROISOQUINOLINE-8-SULFONAMIDE, SULFATE ION | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2015-01-05 | | Release date: | 2016-01-06 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structure of the Saccharomyces cerevisiae Hrr25:Mam1 monopolin subcomplex reveals a novel kinase regulator.
Embo J., 35, 2016
|
|
4XHM
 
 | |
4XHN
 
 | | Bacillus thuringiensis ParM with AMPPNP | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Uncharacterized protein | | Authors: | Jiang, S.M, Robinson, R.C. | | Deposit date: | 2015-01-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A novel plasmid-segregating actin-like protein from Bacillus thuringiensis forms dynamically unstable tubules
to be published
|
|
4XHO
 
 | | Bacillus thuringiensis ParM with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, Uncharacterized protein | | Authors: | Jiang, S.M, Robinson, R.C. | | Deposit date: | 2015-01-06 | | Release date: | 2016-03-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | A novel plasmid-segregating actin-like protein from Bacillus thuringiensis forms dynamically unstable tubules
to be published
|
|
4XHP
 
 | |
4XHQ
 
 | |
4XHR
 
 | | Structure of a phospholipid trafficking complex, native | | Descriptor: | Mitochondrial distribution and morphology protein 35, Protein UPS1, mitochondrial | | Authors: | Yu, F, He, F, Wang, C, Zhang, P. | | Deposit date: | 2015-01-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis of intramitochondrial phosphatidic acid transport mediated by Ups1-Mdm35 complex
Embo Rep., 16, 2015
|
|
4XHS
 
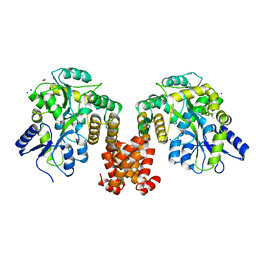 | | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction | | Descriptor: | FORMIC ACID, Maltose-binding periplasmic protein,NACHT, LRR and PYD domains-containing protein 12, ... | | Authors: | Jin, T, Huang, M, Jiang, J, Xiao, T. | | Deposit date: | 2015-01-06 | | Release date: | 2016-01-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of human NLRP12 PYD domain and implication in homotypic interaction
To Be Published
|
|
4XHT
 
 | |
4XHU
 
 | |
4XHV
 
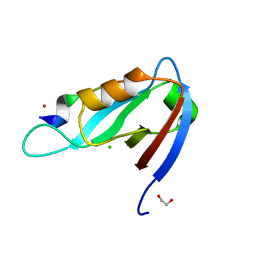 | | Crystal structure of Drosophila Spinophilin-PDZ and a C-terminal peptide of Neurexin | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, LP20995p, ... | | Authors: | Driller, J.H, Muhammad, K.G.H, Reddy, S, Rey, U, Boehme, M.A, Hollmann, C, Ramesh, N, Depner, H, Luetzkendorf, J, Matkovic, T, Bergeron, D, Quentin, C, Schmoranzer, J, Goettfert, F, Holt, M, Wahl, M.C, Hell, S.W, Walter, A, Sigrist, S.J, Loll, B. | | Deposit date: | 2015-01-06 | | Release date: | 2015-07-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Presynaptic spinophilin tunes neurexin signalling to control active zone architecture and function.
Nat Commun, 6, 2015
|
|
4XHW
 
 | |
4XHX
 
 | |
4XHY
 
 | | NADH:FMN oxidoreductase from Paracoccus denitrificans | | Descriptor: | Flavin reductase domain protein, FMN-binding protein | | Authors: | Sedlacek, V, Klumpler, T, Marek, J, Kucera, I. | | Deposit date: | 2015-01-06 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Biochemical properties and crystal structure of the flavin reductase FerA from Paracoccus denitrificans.
Microbiol. Res., 188-189, 2016
|
|
