6U7X
 
 | | NMR solution structure of triazole bridged plasmin inhibitor | | Descriptor: | 1-methyl-1H-1,2,3-triazole, GLY-ARG-ALA-TYR-LYS-SER-LYS-PRO-PRO-ILE-ALA-PHE-PRO-ASP | | Authors: | White, A.M, Harvey, P.J, Wang, C.K, Durek, T, Craik, D.J. | | Deposit date: | 2019-09-03 | | Release date: | 2020-04-22 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Application and Structural Analysis of Triazole-Bridged Disulfide Mimetics in Cyclic Peptides.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6UCH
 
 | | SMARCB1 nucleosome-interacting C-terminal alpha helix | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily B member 1 | | Authors: | Valencia, A.M, Sun, Z.Y.J, Seo, H.S, Vangos, H.S, Yeoh, Z.C, Mashtalir, N, Dhe-Paganon, S, Kadoch, C. | | Deposit date: | 2019-09-16 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recurrent SMARCB1 Mutations Reveal a Nucleosome Acidic Patch Interaction Site That Potentiates mSWI/SNF Complex Chromatin Remodeling.
Cell, 179, 2019
|
|
6UCJ
 
 | | proIAPP in DPC Micelles - Two-Conformer Ensemble Refinement, Open Conformer | | Descriptor: | Islet amyloid polypeptide | | Authors: | DeLisle, C.F, Malooley, A.L, Banerjee, I, Lorieau, J.L. | | Deposit date: | 2019-09-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Pro-islet amyloid polypeptide in micelles contains a helical prohormone segment.
Febs J., 287, 2020
|
|
6UCK
 
 | | proIAPP in DPC Micelles - Two-Conformer Ensemble Refinement, Bent Conformer | | Descriptor: | Islet amyloid polypeptide | | Authors: | DeLisle, C.F, Malooley, A.L, Banerjee, I, Lorieau, J.L. | | Deposit date: | 2019-09-16 | | Release date: | 2020-02-26 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Pro-islet amyloid polypeptide in micelles contains a helical prohormone segment.
Febs J., 287, 2020
|
|
6UCO
 
 | |
6UCP
 
 | |
6UD0
 
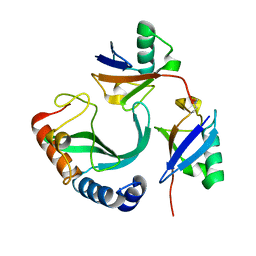 | | Solution-state NMR structural ensemble of human Tsg101 UEV in complex with K63-linked diubiquitin | | Descriptor: | Tumor susceptibility gene 101 protein, Ubiquitin | | Authors: | Strickland, M, Watanabe, S, Bonn, S.M, Camara, C.M, Fushman, D, Carter, C.A, Tjandra, N. | | Deposit date: | 2019-09-18 | | Release date: | 2021-03-17 | | Last modified: | 2024-10-16 | | Method: | SOLUTION NMR | | Cite: | Tsg101/ESCRT-I Recruitment Regulated by the Dual Binding Modes of K63-Linked Diubiquitin
Structure, 2021
|
|
6UF2
 
 | |
6UHW
 
 | |
6UJU
 
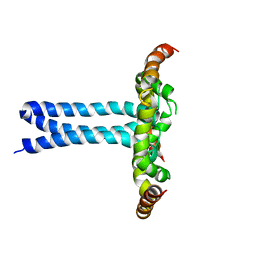 | | Structure of the HIV-1 gp41 transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
6UJV
 
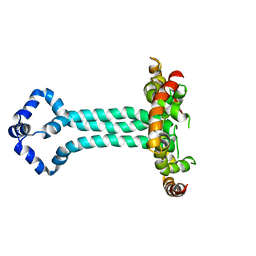 | | Model of the HIV-1 gp41 membrane-proximal external region, transmembrane domain and cytoplasmic tail (LLP2) | | Descriptor: | Envelope glycoprotein GP41 | | Authors: | Piai, A, Fu, Q, Cai, Y, Ghantous, F, Xiao, T, Shaik, M.M, Peng, H, Rits-Volloch, S, Liu, Z, Chen, W, Seaman, M.S, Chen, B, Chou, J.J. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Nat Commun, 11, 2020
|
|
6UM9
 
 | |
6URP
 
 | |
6URS
 
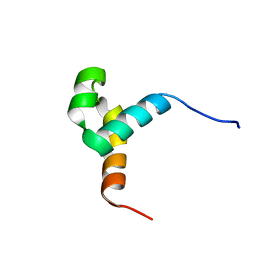 | | Sleeping Beauty transposase PAI subdomain mutant - H19Y | | Descriptor: | Sleeping Beauty transposase PAI subdomain | | Authors: | Nesmelova, I.V, Leighton, G.O, Yan, C, Lustig, J, Corona, R.I, Guo, J.T, Ivics, Z. | | Deposit date: | 2019-10-24 | | Release date: | 2020-10-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | H19Y mutation in the primary DNA-recognition subdomain of the Sleeping Beauty transposase improves structural stability, transposon DNA-binding and transposition
To Be Published
|
|
6UT2
 
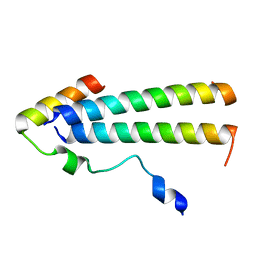 | | 3D structure of the leiomodin/tropomyosin binding interface | | Descriptor: | Leiomodin-2, Tropomyosin alpha-1 chain chimeric peptide | | Authors: | Tolkatchev, D, Smith, G.E, Helms, G.L, Cort, J.R, Kostyukova, A.S. | | Deposit date: | 2019-10-29 | | Release date: | 2020-09-30 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Leiomodin creates a leaky cap at the pointed end of actin-thin filaments.
Plos Biol., 18, 2020
|
|
6UX5
 
 | |
6UXS
 
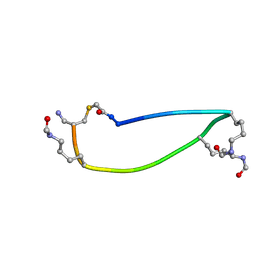 | |
6UYI
 
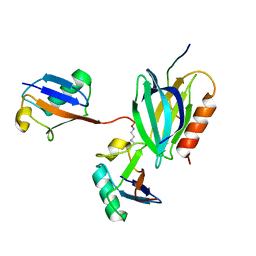 | | hRpn13:hRpn2:K48-diubiquitin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Lu, X, Walters, K.J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-13 | | Method: | SOLUTION NMR | | Cite: | An Extended Conformation for K48 Ubiquitin Chains Revealed by the hRpn2:Rpn13:K48-Diubiquitin Structure.
Structure, 28, 2020
|
|
6UYJ
 
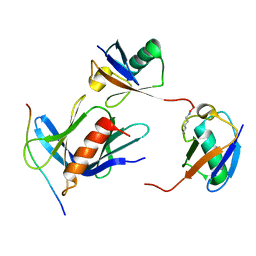 | | hRpn13:hRpn2:K48-diubiquitin | | Descriptor: | 26S proteasome non-ATPase regulatory subunit 1, Proteasomal ubiquitin receptor ADRM1, Ubiquitin | | Authors: | Lu, X, Walters, K.J. | | Deposit date: | 2019-11-13 | | Release date: | 2020-03-11 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | An Extended Conformation for K48 Ubiquitin Chains Revealed by the hRpn2:Rpn13:K48-Diubiquitin Structure.
Structure, 28, 2020
|
|
6UZ4
 
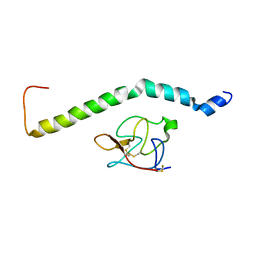 | | Solution structure of AGL55-Kringle 2 complex | | Descriptor: | M protein, Plasminogen | | Authors: | Qiu, C, Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-11-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural evolution of the A-domain in plasminogen-binding Group A streptococcal M-protein reflects improved adaptability of the pathogen to the host
To Be Published
|
|
6UZ5
 
 | | Solution structure of KTI55-Kringle 2 complex | | Descriptor: | M protein, Plasminogen | | Authors: | Qiu, C, Yuan, Y, Castellino, F.J. | | Deposit date: | 2019-11-14 | | Release date: | 2020-12-09 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Structural evolution of the A-domain in plasminogen-binding Group A streptococcal M-protein reflects improved adaptability of the pathogen to the host
To Be Published
|
|
6UZJ
 
 | | NMR structure of the HACS1 SH3 domain | | Descriptor: | SAM domain-containing protein SAMSN-1 | | Authors: | Donaldson, L.W. | | Deposit date: | 2019-11-15 | | Release date: | 2019-11-27 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | HACS1 signaling adaptor protein recognizes a motif in the paired immunoglobulin receptor B cytoplasmic domain.
Commun Biol, 3, 2020
|
|
6V0L
 
 | | PDGFR-b Promoter Forms a G-Vacancy Quadruplex that Can be Complemented by dGMP: Molecular Structure and Recognition of Guanine Derivatives and Metabolites | | Descriptor: | 2'-DEOXYGUANOSINE-5'-MONOPHOSPHATE, DNA (5'-D(*(3D1)P*AP*GP*GP*GP*AP*GP*GP*GP*CP*GP*GP*CP*GP*GP*GP*AP*CP*A)-3') | | Authors: | Wang, K.B, Dickerhoff, J, Wu, G, Yang, D. | | Deposit date: | 2019-11-18 | | Release date: | 2020-03-11 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | PDGFR-beta Promoter Forms a Vacancy G-Quadruplex that Can Be Filled in by dGMP: Solution Structure and Molecular Recognition of Guanine Metabolites and Drugs.
J.Am.Chem.Soc., 142, 2020
|
|
6V1N
 
 | | CSP1-E1A-cyc(Dap6E10) | | Descriptor: | Competence-stimulating peptide type 1 | | Authors: | Yang, Y. | | Deposit date: | 2019-11-20 | | Release date: | 2020-01-08 | | Last modified: | 2025-04-02 | | Method: | SOLUTION NMR | | Cite: | Designing cyclic competence-stimulating peptide (CSP) analogs with pan-group quorum-sensing inhibition activity inStreptococcus pneumoniae.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6V1W
 
 | |
