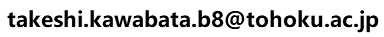 |
The ghecom program read a structure file in PDB format.
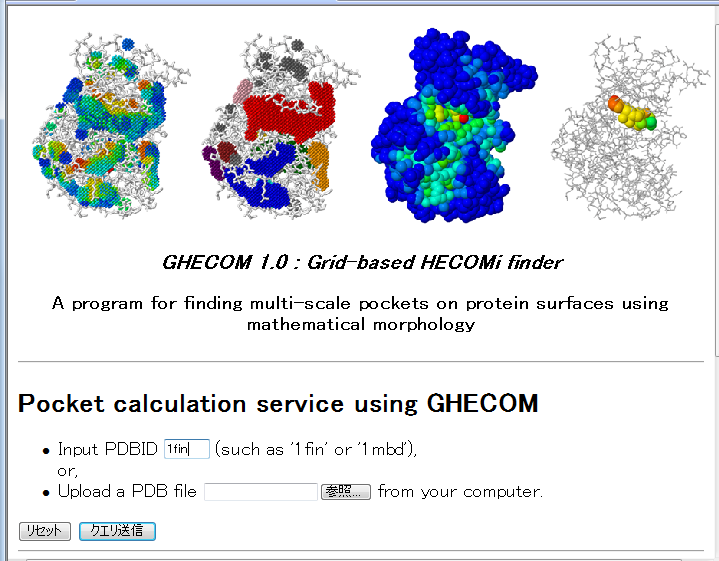
A user can submit a PDB structual file by followint two ways: (1) Input PDB code (such 1fin, 1mbd) (2) Upload PDB file from the user's client computer. After that, click the [submit] button. |
|
(1)A PDB file often contains more than one molecule.
A user has to select "Receptor",Ligand", "ignore" for each molecule. Receptor (required) :receptor proteins, to which ligands will bind. Ligand (not required) :ligand molecules, whose depth(Rinaccess) will be calculated. ignore :the program will ignore them. (2) Maximum radius for the large probe should be specified among 12, 10, 8, 6 and 4 angstom. (3) The ghecom normally output pocket regions as a set of grids. When a user want a set of spheres as pockets, change this selection "off" to "on". Finally, pushing the button [ghecom], then the calculation will begin. |
|
|
During the calculation, "the message "Now calculating, please wait a momment.." will be shown. Estimated calculation time is also shown. |
|
|
After finishing the calculation, three types of links for the result will be shown
at the bottom of the page.
(1) Links to the graphical page using jmol. The tutorial of jmol view of pockets is available. (2) Links to the graph page of residue-based pocketness. The tutorial of graph for residue-based pocketness is available. (3) Various links to raw result files. |