7NX8
 
 | | Crystal structure of the K417T mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, CITRIC ACID, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NX9
 
 | | Crystal structure of the N501Y mutant receptor binding domain of SARS-CoV-2 Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, COVOX-222 Fab heavy chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXA
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 B.1.351 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXB
 
 | | Crystal structure of the receptor binding domain of SARS-CoV-2 P.1 variant Spike glycoprotein in complex with COVOX-222 and EY6A Fabs | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, COVOX-222 Fab heavy chain, COVOX-222 Fab light chain, ... | | Authors: | Zhou, D, Ren, J, Stuart, D. | | Deposit date: | 2021-03-17 | | Release date: | 2021-04-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | Antibody evasion by the P.1 strain of SARS-CoV-2.
Cell, 184, 2021
|
|
7NXC
 
 | |
7L56
 
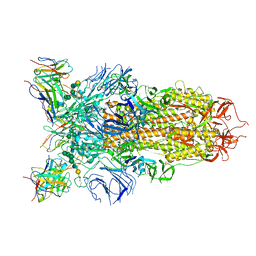 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab 2-43 variable domain heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7L57
 
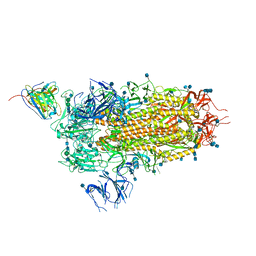 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab 2-15 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (5.87 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7L58
 
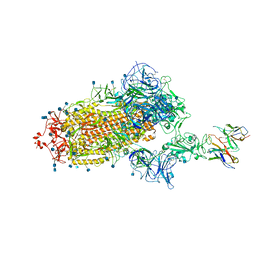 | | Cryo-EM structure of the SARS-CoV-2 spike glycoprotein bound to Fab H4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab H4 variable domain heavy chain, ... | | Authors: | Rapp, M, Shapiro, L. | | Deposit date: | 2020-12-21 | | Release date: | 2021-04-14 | | Last modified: | 2021-04-21 | | Method: | ELECTRON MICROSCOPY (5.07 Å) | | Cite: | Modular basis for potent SARS-CoV-2 neutralization by a prevalent VH1-2-derived antibody class.
Cell Rep, 35, 2021
|
|
7LMC
 
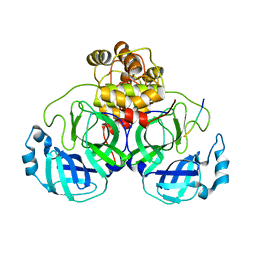 | |
7LXW
 
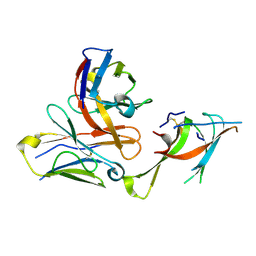 | |
7LXX
 
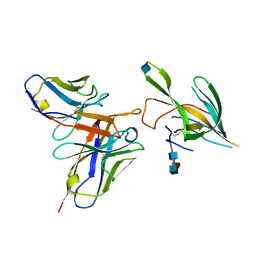 | |
7LXY
 
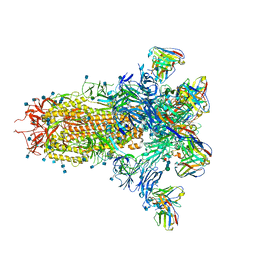 | |
7LXZ
 
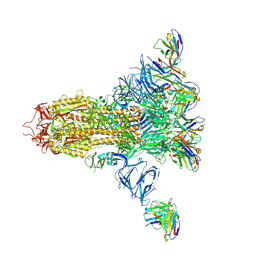 | |
7LY0
 
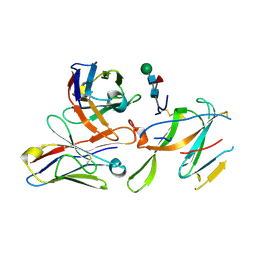 | |
7LY2
 
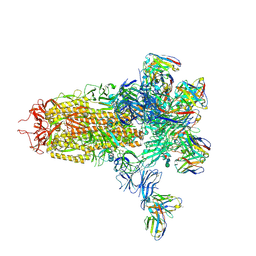 | |
7LY3
 
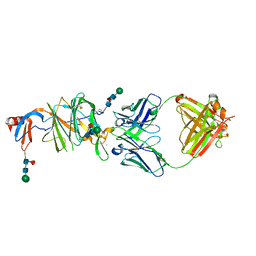 | |
7ME0
 
 | | Cryo-EM structure of SARS-CoV-2 NSP15 NendoU at pH 6.0 | | Descriptor: | Uridylate-specific endoribonuclease | | Authors: | Godoy, A.S, Song, Y, Nakamura, A.M, Noske, G.D, Gawriljuk, V.O, Fernandes, R.S, Oliva, G. | | Deposit date: | 2021-04-06 | | Release date: | 2021-04-14 | | Last modified: | 2023-05-10 | | Method: | ELECTRON MICROSCOPY (2.48 Å) | | Cite: | Allosteric regulation and crystallographic fragment screening of SARS-CoV-2 NSP15 endoribonuclease.
Nucleic Acids Res., 2023
|
|
7D7K
 
 | | The crystal structure of SARS-CoV-2 papain-like protease in apo form | | Descriptor: | 1,2-ETHANEDIOL, CAFFEINE, Non-structural protein 3, ... | | Authors: | Zhao, Y, Sun, L, Yang, H.T, Rao, Z.H. | | Deposit date: | 2020-10-04 | | Release date: | 2021-04-21 | | Last modified: | 2021-11-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | High-throughput screening identifies established drugs as SARS-CoV-2 PLpro inhibitors.
Protein Cell, 12, 2021
|
|
7DX4
 
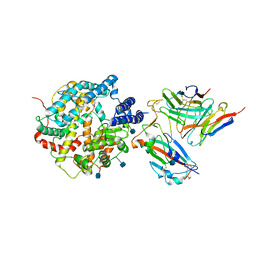 | | The structure of FC08 Fab-hA.CE2-RBD complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, Heavy chain of FC08 Fab, ... | | Authors: | Cao, L, Wang, X. | | Deposit date: | 2021-01-18 | | Release date: | 2021-04-21 | | Last modified: | 2022-02-23 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | A proof of concept for neutralizing antibody-guided vaccine design against SARS-CoV-2.
Natl Sci Rev, 8, 2021
|
|
7KRN
 
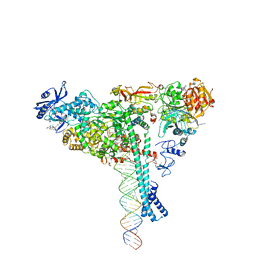 | | Structure of SARS-CoV-2 backtracked complex bound to nsp13 helicase - nsp13(1)-BTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7KRP
 
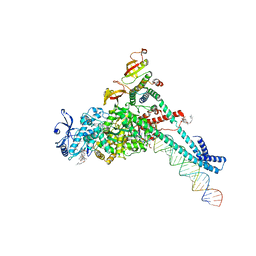 | | Structure of SARS-CoV-2 backtracked complex complex bound to nsp13 helicase - BTC (local refinement) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHAPSO, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2020-11-20 | | Release date: | 2021-04-21 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for backtracking by the SARS-CoV-2 replication-transcription complex.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7LGD
 
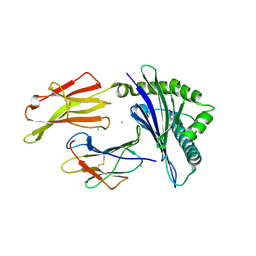 | | HLA-B*07:02 in complex with SARS-CoV-2 nucleocapsid peptide N105-113 | | Descriptor: | Beta-2-microglobulin, CHLORIDE ION, HLA class I histocompatibility antigen, ... | | Authors: | Gras, S, Szeto, C, Chatzileontiadou, D.S.M. | | Deposit date: | 2021-01-20 | | Release date: | 2021-04-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | CD8 + T cells specific for an immunodominant SARS-CoV-2 nucleocapsid epitope cross-react with selective seasonal coronaviruses.
Immunity, 54, 2021
|
|
7B14
 
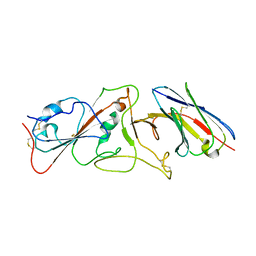 | | Nanobody E bound to Spike-RBD in a localized reconstruction | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2, Spike protein S1 | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-23 | | Release date: | 2021-04-28 | | Last modified: | 2022-12-21 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape
Science, 371, 2021
|
|
7B18
 
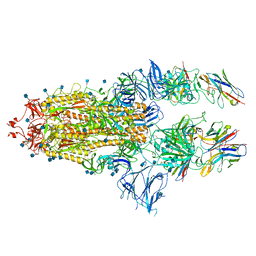 | | SARS-CoV-spike bound to two neutralising nanobodies | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Nanobody against SARS-CoV-2 VHH E, ... | | Authors: | Hallberg, B.M, Das, H. | | Deposit date: | 2020-11-24 | | Release date: | 2021-04-28 | | Method: | ELECTRON MICROSCOPY (2.62 Å) | | Cite: | Structure-guided multivalent nanobodies block SARS-CoV-2 infection and suppress mutational escape.
Science, 371, 2021
|
|
7B62
 
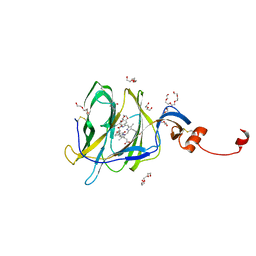 | | Crystal structure of SARS-CoV-2 spike protein N-terminal domain in complex with biliverdin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pye, V.E, Rosa, A, Roustan, C, Cherepanov, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|







