9ART
 
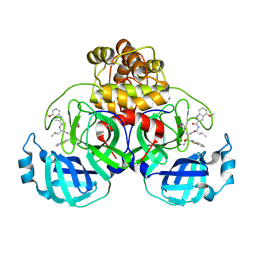 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor 5h | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
8TV7
 
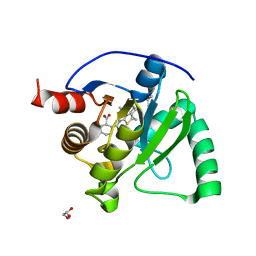 | | SARS-CoV-2 Mac1 in complex with MDOLL-0229 | | Descriptor: | (1R,2R)-2-{[3-(methoxycarbonyl)-4,5,6,7,8,9-hexahydrocycloocta[b]thiophen-2-yl]carbamoyl}cyclohexane-1-carboxylic acid, GLYCEROL, Papain-like protease nsp3 | | Authors: | Wazir, S, Maksimainen, M, Lehtio, L. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Discovery of 2-amide-3-methylester thiophenes inhibiting SARS-CoV-2 ADP-ribosyl hydrolysing macrodomain and coronavirus replication and coronavirus replication
To Be Published
|
|
9ARQ
 
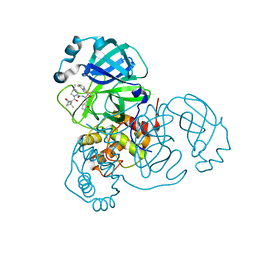 | | Crystal structure of SARS-CoV-2 main protease (authentic protein) in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
8XI6
 
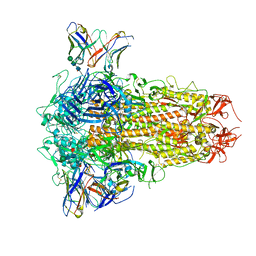 | | SARS-CoV-2 Omicron BQ.1.1 Variant Spike Protein Complexed with MO11 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Ishimaru, H, Nishimura, M, Shigematsu, H, Marini, M.I, Hasegawa, N, Takamiya, R, Iwata, S, Mori, Y. | | Deposit date: | 2023-12-19 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Epitopes of an antibody that neutralizes a wide range of SARS-CoV-2 variants in a conserved subdomain 1 of the spike protein.
J.Virol., 2024
|
|
8UD2
 
 | |
9ARS
 
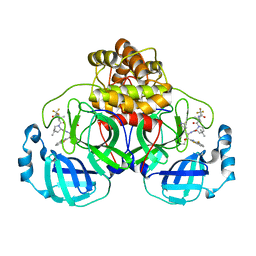 | | Crystal structure of SARS-CoV-2 main protease E166V mutant in complex with an inhibitor TKB-245 | | Descriptor: | (1R,2S,5S)-N-{(1S,2S)-1-(4-fluoro-1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Misumi, S, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
8WMF
 
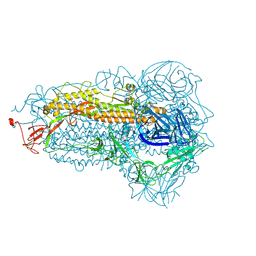 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-1 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-10-03 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.51 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant
Biorxiv, 2024
|
|
9AVQ
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor Nirmatrelvir | | Descriptor: | (1R,2S,5S)-N-{(1E,2S)-1-imino-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5, DI(HYDROXYETHYL)ETHER | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-03-04 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
8UD3
 
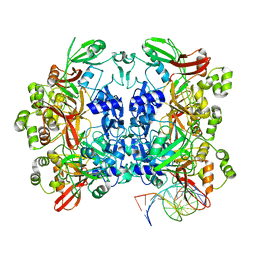 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, consensus form | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
8WMD
 
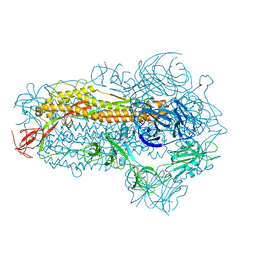 | | Structure of the SARS-CoV-2 EG.5.1 spike glycoprotein (closed-2 state) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Nomai, T, Anraku, Y, Kita, S, Hashiguchi, T, Maenaka, K. | | Deposit date: | 2023-10-03 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.71 Å) | | Cite: | Virological characteristics of the SARS-CoV-2 Omicron EG.5.1 variant
Biorxiv, 2024
|
|
7YLD
 
 | |
8UD5
 
 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 2 | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
8UD4
 
 | | SARS-CoV-2 Nsp15 bound to poly(A/U) RNA, state 1 | | Descriptor: | Non-structural protein 15, RNA (35-MER) | | Authors: | Ito, F, Yang, H, Zhou, Z.H, Chen, X.S. | | Deposit date: | 2023-09-28 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis for polyuridine tract recognition by SARS-CoV-2 Nsp15.
Protein Cell, 2024
|
|
8XEA
 
 | | XBB.1.5 spike protein in complex with BD55-1205 | | Descriptor: | BD55-1205 heavy chain, BD55-1205 light chain, Spike glycoprotein | | Authors: | Feng, L.L. | | Deposit date: | 2023-12-11 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | XBB.1.5 spike protein in complex with BD55-1205
To Be Published
|
|







