+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-7068 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | rsCSP + Fab311 Complex | |||||||||
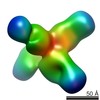
 Map data Map data | rsCSP + Fab311 Complex | |||||||||
 Sample Sample |
| |||||||||
| Biological species |   Plasmodium falciparum (malaria parasite P. falciparum) / Plasmodium falciparum (malaria parasite P. falciparum) /   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  negative staining / Resolution: 25.0 Å negative staining / Resolution: 25.0 Å | |||||||||
 Authors Authors | Torres JL / Ward AB | |||||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2017 Journal: Proc Natl Acad Sci U S A / Year: 2017Title: Structural basis for antibody recognition of the NANP repeats in circumsporozoite protein. Authors: David Oyen / Jonathan L Torres / Ulrike Wille-Reece / Christian F Ockenhouse / Daniel Emerling / Jacob Glanville / Wayne Volkmuth / Yevel Flores-Garcia / Fidel Zavala / Andrew B Ward / C ...Authors: David Oyen / Jonathan L Torres / Ulrike Wille-Reece / Christian F Ockenhouse / Daniel Emerling / Jacob Glanville / Wayne Volkmuth / Yevel Flores-Garcia / Fidel Zavala / Andrew B Ward / C Richter King / Ian A Wilson /  Abstract: Acquired resistance against antimalarial drugs has further increased the need for an effective malaria vaccine. The current leading candidate, RTS,S, is a recombinant circumsporozoite protein (CSP)- ...Acquired resistance against antimalarial drugs has further increased the need for an effective malaria vaccine. The current leading candidate, RTS,S, is a recombinant circumsporozoite protein (CSP)-based vaccine against that contains 19 NANP repeats followed by a thrombospondin repeat domain. Although RTS,S has undergone extensive clinical testing and has progressed through phase III clinical trials, continued efforts are underway to enhance its efficacy and duration of protection. Here, we determined that two monoclonal antibodies (mAbs 311 and 317), isolated from a recent controlled human malaria infection trial exploring a delayed fractional dose, inhibit parasite development in vivo by at least 97%. Crystal structures of antibody fragments (Fabs) 311 and 317 with an (NPNA) peptide illustrate their different binding modes. Notwithstanding, one and three of the three NPNA repeats adopt similar well-defined type I β-turns with Fab311 and Fab317, respectively. Furthermore, to explore antibody binding in the context of CSP, we used negative-stain electron microscopy on a recombinant shortened CSP (rsCSP) construct saturated with Fabs. Both complexes display a compact rsCSP with multiple Fabs bound, with the rsCSP-Fab311 complex forming a highly organized helical structure. Together, these structural insights may aid in the design of a next-generation malaria vaccine. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_7068.map.gz emd_7068.map.gz | 11.8 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-7068-v30.xml emd-7068-v30.xml emd-7068.xml emd-7068.xml | 12.4 KB 12.4 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_7068.png emd_7068.png | 28 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7068 http://ftp.pdbj.org/pub/emdb/structures/EMD-7068 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7068 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7068 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_7068.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_7068.map.gz / Format: CCP4 / Size: 15.6 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | rsCSP + Fab311 Complex | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 2.05 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : Complex containing recombinant, shortened circumsporozite protein...
| Entire | Name: Complex containing recombinant, shortened circumsporozite protein (CSP) and fragment antigen binding (Fab) 311 |
|---|---|
| Components |
|
-Supramolecule #1: Complex containing recombinant, shortened circumsporozite protein...
| Supramolecule | Name: Complex containing recombinant, shortened circumsporozite protein (CSP) and fragment antigen binding (Fab) 311 type: complex / ID: 1 / Parent: 0 Details: Plasmodium falciparum's circumsporozite protein in complex with Fab311, a protecting antibody from the RTS,S clinical trial |
|---|
-Supramolecule #2: Complex containing recombinant, shortened circumsporozite protein...
| Supramolecule | Name: Complex containing recombinant, shortened circumsporozite protein (CSP) type: complex / ID: 2 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Plasmodium falciparum (malaria parasite P. falciparum) Plasmodium falciparum (malaria parasite P. falciparum)Strain: 3D7 |
| Recombinant expression | Organism:   Escherichia coli (E. coli) Escherichia coli (E. coli) |
| Molecular weight | Theoretical: 492 KDa |
-Supramolecule #3: Fragment antigen binding (Fab) 311
| Supramolecule | Name: Fragment antigen binding (Fab) 311 / type: complex / ID: 3 / Parent: 1 |
|---|---|
| Source (natural) | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Recombinant expression | Organism:   Homo sapiens (human) Homo sapiens (human) |
| Molecular weight | Theoretical: 50 KDa |
-Experimental details
-Structure determination
| Method |  negative staining negative staining |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 0.01 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.4 Component:
| |||||||||
| Staining | Type: NEGATIVE / Material: 2% (w/v) Uranyl Formate Details: add 3 uL sample, blot, add 3 uL 2% uranyl formate, blot immediately, add 3 uL 2% uranyl formate, blot after 30 seconds | |||||||||
| Grid | Model: Homemade / Material: COPPER / Mesh: 400 / Support film - Material: CELLULOSE ACETATE / Support film - topology: CONTINUOUS / Pretreatment - Type: GLOW DISCHARGE / Details: 15 mA |
- Electron microscopy
Electron microscopy
| Microscope | FEI TECNAI SPIRIT |
|---|---|
| Electron beam | Acceleration voltage: 120 kV / Electron source: LAB6 |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / Nominal defocus min: 1.5 µm / Nominal magnification: 52000 Bright-field microscopy / Cs: 2.7 mm / Nominal defocus min: 1.5 µm / Nominal magnification: 52000 |
| Sample stage | Specimen holder model: SIDE ENTRY, EUCENTRIC / Cooling holder cryogen: NITROGEN |
| Image recording | Film or detector model: TVIPS TEMCAM-F416 (4k x 4k) / Number grids imaged: 1 / Average electron dose: 25.0 e/Å2 |
| Experimental equipment |  Model: Tecnai Spirit / Image courtesy: FEI Company |
- Image processing
Image processing
| Particle selection | Number selected: 6320 |
|---|---|
| Initial angle assignment | Type: PROJECTION MATCHING Projection matching processing - Number reference projections: 20 Software - Name: EMAN (ver. 2) / Software - details: e2initialmodel.py / Details: Used EMAN2 e2initialmodel |
| Final 3D classification | Number classes: 3 / Avg.num./class: 7000 / Software - Name: RELION (ver. 2.0) |
| Final angle assignment | Type: NOT APPLICABLE |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: RELION (ver. 2.0) / Number images used: 6320 |
 Movie
Movie Controller
Controller