[English] 日本語
 Yorodumi
Yorodumi- PDB-6qm7: Leishmania tarentolae proteasome 20S subunit complexed with GSK3494245 -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6qm7 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Leishmania tarentolae proteasome 20S subunit complexed with GSK3494245 | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  HYDROLASE / Proteasome 20S subunit HYDROLASE / Proteasome 20S subunit | ||||||
| Function / homology |  Function and homology information Function and homology informationproteasome core complex /  proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / proteasome core complex, alpha-subunit complex / threonine-type endopeptidase activity / proteolysis involved in protein catabolic process / proteasomal protein catabolic process / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process / proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / proteasome core complex, alpha-subunit complex / threonine-type endopeptidase activity / proteolysis involved in protein catabolic process / proteasomal protein catabolic process / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process /  hydrolase activity ...proteasome core complex / hydrolase activity ...proteasome core complex /  proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / proteasome core complex, alpha-subunit complex / threonine-type endopeptidase activity / proteolysis involved in protein catabolic process / proteasomal protein catabolic process / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process / proteasome endopeptidase complex / proteasome core complex, beta-subunit complex / proteasome core complex, alpha-subunit complex / threonine-type endopeptidase activity / proteolysis involved in protein catabolic process / proteasomal protein catabolic process / ubiquitin-dependent protein catabolic process / proteasome-mediated ubiquitin-dependent protein catabolic process /  hydrolase activity / hydrolase activity /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |  Leishmania tarentolae (eukaryote) Leishmania tarentolae (eukaryote) | ||||||
| Method |  ELECTRON MICROSCOPY / ELECTRON MICROSCOPY /  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 2.8 Å cryo EM / Resolution: 2.8 Å | ||||||
 Authors Authors | Rowland, P. / Goswami, P. | ||||||
 Citation Citation |  Journal: Proc Natl Acad Sci U S A / Year: 2019 Journal: Proc Natl Acad Sci U S A / Year: 2019Title: Preclinical candidate for the treatment of visceral leishmaniasis that acts through proteasome inhibition. Authors: Susan Wyllie / Stephen Brand / Michael Thomas / Manu De Rycker / Chun-Wa Chung / Imanol Pena / Ryan P Bingham / Juan A Bueren-Calabuig / Juan Cantizani / David Cebrian / Peter D Craggs / ...Authors: Susan Wyllie / Stephen Brand / Michael Thomas / Manu De Rycker / Chun-Wa Chung / Imanol Pena / Ryan P Bingham / Juan A Bueren-Calabuig / Juan Cantizani / David Cebrian / Peter D Craggs / Liam Ferguson / Panchali Goswami / Judith Hobrath / Jonathan Howe / Laura Jeacock / Eun-Jung Ko / Justyna Korczynska / Lorna MacLean / Sujatha Manthri / Maria S Martinez / Lydia Mata-Cantero / Sonia Moniz / Andrea Nühs / Maria Osuna-Cabello / Erika Pinto / Jennifer Riley / Sharon Robinson / Paul Rowland / Frederick R C Simeons / Yoko Shishikura / Daniel Spinks / Laste Stojanovski / John Thomas / Stephen Thompson / Elisabet Viayna Gaza / Richard J Wall / Fabio Zuccotto / David Horn / Michael A J Ferguson / Alan H Fairlamb / Jose M Fiandor / Julio Martin / David W Gray / Timothy J Miles / Ian H Gilbert / Kevin D Read / Maria Marco / Paul G Wyatt /   Abstract: Visceral leishmaniasis (VL), caused by the protozoan parasites and , is one of the major parasitic diseases worldwide. There is an urgent need for new drugs to treat VL, because current therapies ...Visceral leishmaniasis (VL), caused by the protozoan parasites and , is one of the major parasitic diseases worldwide. There is an urgent need for new drugs to treat VL, because current therapies are unfit for purpose in a resource-poor setting. Here, we describe the development of a preclinical drug candidate, GSK3494245/DDD01305143/compound 8, with potential to treat this neglected tropical disease. The compound series was discovered by repurposing hits from a screen against the related parasite Subsequent optimization of the chemical series resulted in the development of a potent cidal compound with activity against a range of clinically relevant and isolates. Compound 8 demonstrates promising pharmacokinetic properties and impressive in vivo efficacy in our mouse model of infection comparable with those of the current oral antileishmanial miltefosine. Detailed mode of action studies confirm that this compound acts principally by inhibition of the chymotrypsin-like activity catalyzed by the β5 subunit of the proteasome. High-resolution cryo-EM structures of apo and compound 8-bound 20S proteasome reveal a previously undiscovered inhibitor site that lies between the β4 and β5 proteasome subunits. This induced pocket exploits β4 residues that are divergent between humans and kinetoplastid parasites and is consistent with all of our experimental and mutagenesis data. As a result of these comprehensive studies and due to a favorable developability and safety profile, compound 8 is being advanced toward human clinical trials. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6qm7.cif.gz 6qm7.cif.gz | 1.2 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6qm7.ent.gz pdb6qm7.ent.gz | 1012.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6qm7.json.gz 6qm7.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/qm/6qm7 https://data.pdbj.org/pub/pdb/validation_reports/qm/6qm7 ftp://data.pdbj.org/pub/pdb/validation_reports/qm/6qm7 ftp://data.pdbj.org/pub/pdb/validation_reports/qm/6qm7 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  4590MC  4591C  6qm8C M: map data used to model this data C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments:
NCS ensembles :
NCS oper:
|
- Components
Components
-Protein , 14 types, 28 molecules AOBPCQDRESFTGUHVIWJXKYLZMaNb
| #1: Protein | Mass: 27178.107 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: Q7JMX2*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: Q7JMX2*PLUS#2: Protein | Mass: 25179.559 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: Q9GNZ8*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: Q9GNZ8*PLUS#3: Protein | Mass: 32321.438 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A4HVX3*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A4HVX3*PLUS#4: Protein | Mass: 27821.605 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A4HUT6*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A4HUT6*PLUS#5: Protein | Mass: 38312.316 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A4HZI9*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A4HZI9*PLUS#6: Protein | Mass: 47978.633 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A4IDD0*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A4IDD0*PLUS#7: Protein | Mass: 25591.826 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A4I357*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A4I357*PLUS#8: Protein | Mass: 30280.010 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A4HV47*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A4HV47*PLUS#9: Protein | Mass: 27603.570 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A4HN55*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A4HN55*PLUS#10: Protein | Mass: 22470.887 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A4I384*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A4I384*PLUS#11: Protein | Mass: 23065.291 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A4ICV5*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A4ICV5*PLUS#12: Protein | Mass: 33704.867 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A4IDD6*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A4IDD6*PLUS#13: Protein | Mass: 37676.910 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A4HSQ5*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A4HSQ5*PLUS#14: Protein | Mass: 24737.232 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)  Leishmania tarentolae (eukaryote) / References: UniProt: A0A381MU64*PLUS Leishmania tarentolae (eukaryote) / References: UniProt: A0A381MU64*PLUS |
|---|
-Non-polymers , 2 types, 336 molecules 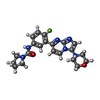


| #15: Chemical | | #16: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  ELECTRON MICROSCOPY ELECTRON MICROSCOPY |
|---|---|
| EM experiment | Aggregation state: PARTICLE / 3D reconstruction method:  single particle reconstruction single particle reconstruction |
- Sample preparation
Sample preparation
| Component | Name: Proteasome 20S subunit / Type: COMPLEX / Entity ID: #1-#14 / Source: NATURAL |
|---|---|
| Source (natural) | Organism:  Leishmania tarentolae (eukaryote) Leishmania tarentolae (eukaryote) |
| Buffer solution | pH: 7.5 |
| Specimen | Embedding applied: NO / Shadowing applied: NO / Staining applied : NO / Vitrification applied : NO / Vitrification applied : YES : YES |
Vitrification | Cryogen name: ETHANE |
- Electron microscopy imaging
Electron microscopy imaging
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
|---|---|
| Microscopy | Model: FEI TITAN KRIOS |
| Electron gun | Electron source : :  FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM FIELD EMISSION GUN / Accelerating voltage: 300 kV / Illumination mode: FLOOD BEAM |
| Electron lens | Mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Image recording | Electron dose: 30 e/Å2 / Detector mode: COUNTING / Film or detector model: FEI FALCON III (4k x 4k) |
- Processing
Processing
| Software | Name: REFMAC / Version: 5.8.0238 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EM software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
CTF correction | Type: PHASE FLIPPING AND AMPLITUDE CORRECTION | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Point symmetry : C1 (asymmetric) : C1 (asymmetric) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
3D reconstruction | Resolution: 2.8 Å / Resolution method: FSC 0.143 CUT-OFF / Num. of particles: 182775 / Symmetry type: POINT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | Protocol: FLEXIBLE FIT | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Atomic model building | PDB-ID: 4R3O | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | Resolution: 2.8→200.09 Å / Cor.coef. Fo:Fc: 0.893 / SU B: 10.057 / SU ML: 0.179 / ESU R: 0.363 Stereochemistry target values: MAXIMUM LIKELIHOOD WITH PHASES Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å / Solvent model: MASK | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 39.918 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Total: 49518 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller






















 PDBj
PDBj




