[English] 日本語
 Yorodumi
Yorodumi- PDB-6mcf: Solution structure of 7SK stem-loop 1 with HIV-1 Tat RNA Binding ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6mcf | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Solution structure of 7SK stem-loop 1 with HIV-1 Tat RNA Binding Domain | |||||||||
 Components Components |
| |||||||||
 Keywords Keywords |  TRANSCRIPTION / RNA Binding Domain / TRANSCRIPTION / RNA Binding Domain /  RNA BINDING PROTEIN RNA BINDING PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology information trans-activation response element binding / Interactions of Tat with host cellular proteins / protein serine/threonine phosphatase inhibitor activity / positive regulation of viral transcription / modulation by virus of host chromatin organization / symbiont-mediated suppression of host translation initiation / molecular sequestering activity / evasion of host immune response / host cell nucleolus / trans-activation response element binding / Interactions of Tat with host cellular proteins / protein serine/threonine phosphatase inhibitor activity / positive regulation of viral transcription / modulation by virus of host chromatin organization / symbiont-mediated suppression of host translation initiation / molecular sequestering activity / evasion of host immune response / host cell nucleolus /  actinin binding ... actinin binding ... trans-activation response element binding / Interactions of Tat with host cellular proteins / protein serine/threonine phosphatase inhibitor activity / positive regulation of viral transcription / modulation by virus of host chromatin organization / symbiont-mediated suppression of host translation initiation / molecular sequestering activity / evasion of host immune response / host cell nucleolus / trans-activation response element binding / Interactions of Tat with host cellular proteins / protein serine/threonine phosphatase inhibitor activity / positive regulation of viral transcription / modulation by virus of host chromatin organization / symbiont-mediated suppression of host translation initiation / molecular sequestering activity / evasion of host immune response / host cell nucleolus /  actinin binding / negative regulation of peptidyl-threonine phosphorylation / Pausing and recovery of Tat-mediated HIV elongation / Tat-mediated HIV elongation arrest and recovery / RNA-binding transcription regulator activity / Tat-mediated elongation of the HIV-1 transcript / Formation of HIV-1 elongation complex containing HIV-1 Tat / actinin binding / negative regulation of peptidyl-threonine phosphorylation / Pausing and recovery of Tat-mediated HIV elongation / Tat-mediated HIV elongation arrest and recovery / RNA-binding transcription regulator activity / Tat-mediated elongation of the HIV-1 transcript / Formation of HIV-1 elongation complex containing HIV-1 Tat /  cyclin binding / positive regulation of transcription elongation by RNA polymerase II / PKR-mediated signaling / host cell cytoplasm / protein domain specific binding / DNA-templated transcription / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / host cell nucleus / extracellular region / cyclin binding / positive regulation of transcription elongation by RNA polymerase II / PKR-mediated signaling / host cell cytoplasm / protein domain specific binding / DNA-templated transcription / symbiont-mediated suppression of host type I interferon-mediated signaling pathway / host cell nucleus / extracellular region /  metal ion binding metal ion bindingSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human)  Human immunodeficiency virus type 1 group M subtype B Human immunodeficiency virus type 1 group M subtype B | |||||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing simulated annealing | |||||||||
 Authors Authors | Pham, V.V. / D'Souza, V.M. | |||||||||
| Funding support |  United States, 2items United States, 2items
| |||||||||
 Citation Citation |  Journal: Nat Commun / Year: 2018 Journal: Nat Commun / Year: 2018Title: HIV-1 Tat interactions with cellular 7SK and viral TAR RNAs identifies dual structural mimicry. Authors: Pham, V.V. / Salguero, C. / Khan, S.N. / Meagher, J.L. / Brown, W.C. / Humbert, N. / de Rocquigny, H. / Smith, J.L. / D'Souza, V.M. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6mcf.cif.gz 6mcf.cif.gz | 422.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6mcf.ent.gz pdb6mcf.ent.gz | 363.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6mcf.json.gz 6mcf.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mc/6mcf https://data.pdbj.org/pub/pdb/validation_reports/mc/6mcf ftp://data.pdbj.org/pub/pdb/validation_reports/mc/6mcf ftp://data.pdbj.org/pub/pdb/validation_reports/mc/6mcf | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  6mceC  6mciC C: citing same article ( |
|---|---|
| Similar structure data | |
| Other databases |
|
- Links
Links
- Assembly
Assembly
| Deposited unit | 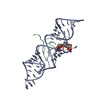
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 18358.910 Da / Num. of mol.: 1 / Source method: obtained synthetically / Source: (synth.)   Homo sapiens (human) Homo sapiens (human) |
|---|---|
| #2: Protein/peptide | Mass: 2162.533 Da / Num. of mol.: 1 / Fragment: RNA BINDING DOMAIN RESIDUES 44-60 / Source method: obtained synthetically Source: (synth.)   Human immunodeficiency virus type 1 group M subtype B Human immunodeficiency virus type 1 group M subtype BReferences: UniProt: P04608 |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sample conditions | Ionic strength: 10mM phosphates, 70mM NaCl mM / Label: 7SK_conditions / pH: 5.4 / Pressure: 100000 Pa / Temperature: 298 K |
-NMR measurement
| NMR spectrometer | Type: Bruker DMX / Manufacturer: Bruker / Model : DMX / Field strength: 700 MHz : DMX / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing / Software ordinal: 6 simulated annealing / Software ordinal: 6 | ||||||||||||||||||||||||
| NMR representative | Selection criteria: fewest violations | ||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the least restraint violations Conformers calculated total number: 20 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller










 PDBj
PDBj






























