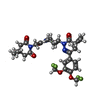
Entry Database : PDB / ID : 6fe3Title Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-1439 Phosphodiesterase Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Trypanosoma brucei (eukaryote)Method / / / Resolution : 1.62 Å Authors Singh, A.K. / Brown, D.G. Journal : To be published Title : TbrPDEB1 structure with inhibitor NPD-1439Authors : Salado, I.G. / Moreno, C. / Sakaine, G. / Singh, A.K. / Blaazer, A.R. / Siderius, M. / Matheeussen, A. / Gul, S. / Maes, L. / Leurs, R. / Brown, D.G. / Augustyns, K. History Deposition Dec 28, 2017 Deposition site / Processing site Revision 1.0 Apr 10, 2019 Provider / Type
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components
 Keywords
Keywords HYDROLASE / Parasitic phosphodiesterase /
HYDROLASE / Parasitic phosphodiesterase /  African trypanosomiasis /
African trypanosomiasis /  sleeping sickness
sleeping sickness Function and homology information
Function and homology information Hydrolases; Acting on ester bonds; Phosphoric-diester hydrolases /
Hydrolases; Acting on ester bonds; Phosphoric-diester hydrolases /  axoneme /
axoneme /  3',5'-cyclic-nucleotide phosphodiesterase activity /
3',5'-cyclic-nucleotide phosphodiesterase activity /  3',5'-cyclic-AMP phosphodiesterase activity / cell morphogenesis /
3',5'-cyclic-AMP phosphodiesterase activity / cell morphogenesis /  signal transduction /
signal transduction /  metal ion binding /
metal ion binding /  cytoplasm
cytoplasm
 Trypanosoma brucei (eukaryote)
Trypanosoma brucei (eukaryote) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  FOURIER SYNTHESIS / Resolution: 1.62 Å
FOURIER SYNTHESIS / Resolution: 1.62 Å  Authors
Authors Citation
Citation Journal: To be published
Journal: To be published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 6fe3.cif.gz
6fe3.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb6fe3.ent.gz
pdb6fe3.ent.gz PDB format
PDB format 6fe3.json.gz
6fe3.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/fe/6fe3
https://data.pdbj.org/pub/pdb/validation_reports/fe/6fe3 ftp://data.pdbj.org/pub/pdb/validation_reports/fe/6fe3
ftp://data.pdbj.org/pub/pdb/validation_reports/fe/6fe3 Links
Links Assembly
Assembly


 Components
Components

 Trypanosoma brucei (eukaryote) / Gene: PDEB1 / Plasmid: pET28a / Production host:
Trypanosoma brucei (eukaryote) / Gene: PDEB1 / Plasmid: pET28a / Production host: 
 Escherichia coli BL21(DE3) (bacteria)
Escherichia coli BL21(DE3) (bacteria) Hydrolases; Acting on ester bonds; Phosphoric-diester hydrolases
Hydrolases; Acting on ester bonds; Phosphoric-diester hydrolases











 Guanidine
Guanidine Formic acid
Formic acid Glycerol
Glycerol Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I03 / Wavelength: 0.97625 Å
/ Beamline: I03 / Wavelength: 0.97625 Å : 0.97625 Å / Relative weight: 1
: 0.97625 Å / Relative weight: 1  Processing
Processing :
:  FOURIER SYNTHESIS
FOURIER SYNTHESIS Movie
Movie Controller
Controller

















 PDBj
PDBj






