+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 6ecj | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Human cytochrome c G41T | |||||||||
 Components Components | Cytochrome c | |||||||||
 Keywords Keywords |  APOPTOSIS / APOPTOSIS /  cytochrome c / cytochrome c /  heme / glycine to threonine substitution heme / glycine to threonine substitution | |||||||||
| Function / homology |  Function and homology information Function and homology informationFormation of apoptosome /  apoptosome / activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c / Release of apoptotic factors from the mitochondria / Respiratory electron transport / apoptosome / activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c / Release of apoptotic factors from the mitochondria / Respiratory electron transport /  cellular respiration / Regulation of the apoptosome activity / Activation of caspases through apoptosome-mediated cleavage / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes ...Formation of apoptosome / cellular respiration / Regulation of the apoptosome activity / Activation of caspases through apoptosome-mediated cleavage / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes ...Formation of apoptosome /  apoptosome / activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c / Release of apoptotic factors from the mitochondria / Respiratory electron transport / apoptosome / activation of cysteine-type endopeptidase activity involved in apoptotic process by cytochrome c / Release of apoptotic factors from the mitochondria / Respiratory electron transport /  cellular respiration / Regulation of the apoptosome activity / Activation of caspases through apoptosome-mediated cleavage / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes / mitochondrial electron transport, cytochrome c to oxygen / mitochondrial electron transport, ubiquinol to cytochrome c / Detoxification of Reactive Oxygen Species / cellular respiration / Regulation of the apoptosome activity / Activation of caspases through apoptosome-mediated cleavage / SMAC (DIABLO) binds to IAPs / SMAC(DIABLO)-mediated dissociation of IAP:caspase complexes / mitochondrial electron transport, cytochrome c to oxygen / mitochondrial electron transport, ubiquinol to cytochrome c / Detoxification of Reactive Oxygen Species /  Pyroptosis / Pyroptosis /  respirasome / intrinsic apoptotic signaling pathway / TP53 Regulates Metabolic Genes / Transcriptional activation of mitochondrial biogenesis / respirasome / intrinsic apoptotic signaling pathway / TP53 Regulates Metabolic Genes / Transcriptional activation of mitochondrial biogenesis /  mitochondrial intermembrane space / Cytoprotection by HMOX1 / mitochondrial intermembrane space / Cytoprotection by HMOX1 /  mitochondrial inner membrane / mitochondrial inner membrane /  electron transfer activity / electron transfer activity /  heme binding / heme binding /  mitochondrion / mitochondrion /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytosol cytosolSimilarity search - Function | |||||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | |||||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.7 Å molecular replacement / Resolution: 2.7 Å | |||||||||
| Model details | G41T variant | |||||||||
 Authors Authors | Fellner, M. / Jameson, G.N.L. / Ledgerwood, E.C. / Wilbanks, S.M. | |||||||||
 Citation Citation |  Journal: Biochem.J. / Year: 2021 Journal: Biochem.J. / Year: 2021Title: Altered structure and dynamics of pathogenic cytochrome c variants correlate with increased apoptotic activity. Authors: Fellner, M. / Parakra, R. / McDonald, K.O. / Kass, I. / Jameson, G.N.L. / Wilbanks, S.M. / Ledgerwood, E.C. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  6ecj.cif.gz 6ecj.cif.gz | 182.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb6ecj.ent.gz pdb6ecj.ent.gz | 144.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  6ecj.json.gz 6ecj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ec/6ecj https://data.pdbj.org/pub/pdb/validation_reports/ec/6ecj ftp://data.pdbj.org/pub/pdb/validation_reports/ec/6ecj ftp://data.pdbj.org/pub/pdb/validation_reports/ec/6ecj | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5exqC  3nwvS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 4 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 5 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 6 | 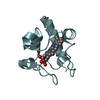
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 7 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 8 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments:
|
 Movie
Movie Controller
Controller







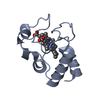


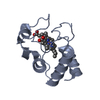


 PDBj
PDBj



















