[English] 日本語
 Yorodumi
Yorodumi- PDB-5nwx: Insight into the molecular recognition mechanism of the coactivat... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5nwx | ||||||
|---|---|---|---|---|---|---|---|
| Title | Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6 | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  TRANSCRIPTION / NcoA1 / TRANSCRIPTION / NcoA1 /  STAT6 / PAS-B domain / STAT6 / PAS-B domain /  transactivation domain transactivation domain | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of mast cell proliferation / NR1H2 & NR1H3 regulate gene expression to control bile acid homeostasis / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / mammary gland morphogenesis / SUMOylation of transcription cofactors / Recycling of bile acids and salts / Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol / Synthesis of bile acids and bile salts via 27-hydroxycholesterol / Synthesis of bile acids and bile salts / cellular response to reactive nitrogen species ...regulation of mast cell proliferation / NR1H2 & NR1H3 regulate gene expression to control bile acid homeostasis / NR1H3 & NR1H2 regulate gene expression linked to cholesterol transport and efflux / mammary gland morphogenesis / SUMOylation of transcription cofactors / Recycling of bile acids and salts / Synthesis of bile acids and bile salts via 7alpha-hydroxycholesterol / Synthesis of bile acids and bile salts via 27-hydroxycholesterol / Synthesis of bile acids and bile salts / cellular response to reactive nitrogen species / negative regulation of type 2 immune response / positive regulation of isotype switching to IgE isotypes / Endogenous sterols / HATs acetylate histones / T-helper 1 cell lineage commitment / STAT6-mediated induction of chemokines / Regulation of lipid metabolism by PPARalpha / isotype switching to IgE isotypes / Cytoprotection by HMOX1 / interleukin-4-mediated signaling pathway / Estrogen-dependent gene expression / nuclear retinoic acid receptor binding / labyrinthine layer morphogenesis / regulation of thyroid hormone mediated signaling pathway / positive regulation of transcription from RNA polymerase II promoter by galactose / positive regulation of female receptivity / mammary gland epithelial cell proliferation / hypothalamus development / male mating behavior / cell surface receptor signaling pathway via JAK-STAT /  estrous cycle / cellular response to Thyroglobulin triiodothyronine / growth hormone receptor signaling pathway via JAK-STAT / nuclear retinoid X receptor binding / response to retinoic acid / estrous cycle / cellular response to Thyroglobulin triiodothyronine / growth hormone receptor signaling pathway via JAK-STAT / nuclear retinoid X receptor binding / response to retinoic acid /  histone acetyltransferase activity / regulation of cellular response to insulin stimulus / histone acetyltransferase activity / regulation of cellular response to insulin stimulus /  histone acetyltransferase / cellular response to hormone stimulus / positive regulation of adipose tissue development / peroxisome proliferator activated receptor signaling pathway / positive regulation of neuron differentiation / histone acetyltransferase / cellular response to hormone stimulus / positive regulation of adipose tissue development / peroxisome proliferator activated receptor signaling pathway / positive regulation of neuron differentiation /  lactation / cerebellum development / Downstream signal transduction / nuclear receptor coactivator activity / response to progesterone / nuclear estrogen receptor binding / lactation / cerebellum development / Downstream signal transduction / nuclear receptor coactivator activity / response to progesterone / nuclear estrogen receptor binding /  nuclear receptor binding / hippocampus development / RNA polymerase II transcription regulatory region sequence-specific DNA binding / mRNA transcription by RNA polymerase II / defense response / nuclear receptor binding / hippocampus development / RNA polymerase II transcription regulatory region sequence-specific DNA binding / mRNA transcription by RNA polymerase II / defense response /  transcription coactivator binding / cerebral cortex development / response to peptide hormone / cellular response to hydrogen peroxide / cytokine-mediated signaling pathway / RNA polymerase II transcription regulator complex / male gonad development / response to estradiol / positive regulation of cold-induced thermogenesis / regulation of cell population proliferation / DNA-binding transcription activator activity, RNA polymerase II-specific / transcription coactivator binding / cerebral cortex development / response to peptide hormone / cellular response to hydrogen peroxide / cytokine-mediated signaling pathway / RNA polymerase II transcription regulator complex / male gonad development / response to estradiol / positive regulation of cold-induced thermogenesis / regulation of cell population proliferation / DNA-binding transcription activator activity, RNA polymerase II-specific /  protein phosphatase binding / Interleukin-4 and Interleukin-13 signaling / protein phosphatase binding / Interleukin-4 and Interleukin-13 signaling /  transcription regulator complex / transcription regulator complex /  transcription coactivator activity / transcription coactivator activity /  protein dimerization activity / DNA-binding transcription factor activity, RNA polymerase II-specific / positive regulation of apoptotic process / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / protein dimerization activity / DNA-binding transcription factor activity, RNA polymerase II-specific / positive regulation of apoptotic process / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity /  chromatin binding / chromatin binding /  chromatin / protein-containing complex binding / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / chromatin / protein-containing complex binding / regulation of transcription by RNA polymerase II / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II /  signal transduction / positive regulation of transcription by RNA polymerase II / protein-containing complex / signal transduction / positive regulation of transcription by RNA polymerase II / protein-containing complex /  DNA binding / DNA binding /  nucleoplasm / identical protein binding / nucleoplasm / identical protein binding /  nucleus / nucleus /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse)  Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.51 Å MOLECULAR REPLACEMENT / Resolution: 2.51 Å | ||||||
 Authors Authors | Russo, L. / Giller, K. / Pfitzner, E. / Griesinger, C. / Becker, S. | ||||||
 Citation Citation |  Journal: Sci Rep / Year: 2017 Journal: Sci Rep / Year: 2017Title: Insight into the molecular recognition mechanism of the coactivator NCoA1 by STAT6. Authors: Russo, L. / Giller, K. / Pfitzner, E. / Griesinger, C. / Becker, S. #1:  Journal: Journal of Molecular Biology / Year: 2004 Journal: Journal of Molecular Biology / Year: 2004Title: Structure of the NCoA-1/SRC-1 PAS-B Domain Bound to the LXXLL Motif of the STAT6 Transactivation domain Authors: Razeto, A. / Ramakrishnan, V. / Litterst, C.M. / Giller, K. / Griesinger, C. / Carlomagno, T. / Lakomek, N. / Heimburg, T. / Lodrini, M. / Pfitzner, E. / Becker, S. #2: Journal: Acta Crystallographica Section D / Year: 2004 Title: Crystallization and preliminary crystallographic studies of the NCoA-1/SRC-1 PAS-B domain bound to the LXXLL motif of the STAT6 transactivation domain Authors: Razeto, A. / Pfitzner, E. / Becker, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5nwx.cif.gz 5nwx.cif.gz | 37.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5nwx.ent.gz pdb5nwx.ent.gz | 24 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5nwx.json.gz 5nwx.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/nw/5nwx https://data.pdbj.org/pub/pdb/validation_reports/nw/5nwx ftp://data.pdbj.org/pub/pdb/validation_reports/nw/5nwx ftp://data.pdbj.org/pub/pdb/validation_reports/nw/5nwx | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  5nwmC 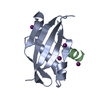 1oj5S C: citing same article ( S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  / NCoA-1 / Nuclear receptor coactivator protein 1 / mNRC-1 / Steroid receptor coactivator 1 / SRC-1 / NCoA-1 / Nuclear receptor coactivator protein 1 / mNRC-1 / Steroid receptor coactivator 1 / SRC-1Mass: 14811.774 Da / Num. of mol.: 1 / Fragment: UNP residues 257-385 / Mutation: K343R Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Gene: Ncoa1, Src1 / Production host: Mus musculus (house mouse) / Gene: Ncoa1, Src1 / Production host:   Escherichia coli BL21(DE3) (bacteria) / References: UniProt: P70365, Escherichia coli BL21(DE3) (bacteria) / References: UniProt: P70365,  histone acetyltransferase histone acetyltransferase |
|---|---|
| #2: Protein/peptide | Mass: 3452.837 Da / Num. of mol.: 1 / Fragment: UNP residues 783-814 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: STAT6 / Production host: synthetic construct (others) / References: UniProt: P42226 Homo sapiens (human) / Gene: STAT6 / Production host: synthetic construct (others) / References: UniProt: P42226 |
| #3: Water | ChemComp-HOH /  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.25 Å3/Da / Density % sol: 44.09 % |
|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion, hanging drop / pH: 6.5 Details: 0.2 M sodium acetate, 0.1 M sodium cacodylate, pH 6.5, 30% PEG 8000, 5% PEG 400 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X10SA / Wavelength: 0.7 Å / Beamline: X10SA / Wavelength: 0.7 Å |
| Detector | Type: MARMOSAIC 225 mm CCD / Detector: CCD / Date: Nov 27, 2010 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.7 Å / Relative weight: 1 : 0.7 Å / Relative weight: 1 |
| Reflection | Resolution: 2.51→53.37 Å / Num. obs: 5471 / % possible obs: 99.8 % / Redundancy: 20.51 % / Rrim(I) all: 0.0246 / Net I/σ(I): 30.26 |
| Reflection shell | Resolution: 2.51→2.61 Å / Redundancy: 19.92 % / Mean I/σ(I) all: 6.08 / Rrim(I) all: 0.1315 / % possible all: 98.3 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: 1OJ5 Resolution: 2.51→53.37 Å / Cor.coef. Fo:Fc: 0.958 / Cor.coef. Fo:Fc free: 0.93 / SU B: 9.917 / SU ML: 0.208 / Cross valid method: THROUGHOUT / ESU R: 0.341 / ESU R Free: 0.248 / Stereochemistry target values: Maximum likelihood / Details: HYDROGENS HAVE BEEN USED IF PRESENT IN THE INPUT
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Ion probe radii: 0.8 Å / Shrinkage radii: 0.8 Å / VDW probe radii: 1.2 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 46.36 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: 1 / Resolution: 2.51→53.37 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
|
 Movie
Movie Controller
Controller











 PDBj
PDBj


















