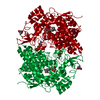
Entry Database : PDB / ID : 5ikrTitle The Structure of Mefenamic Acid Bound to Human Cyclooxygenase-2 Prostaglandin G/H synthase 2 Keywords / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / Resolution : 2.342 Å Authors Orlando, B.J. / Malkowski, M.G. Funding support Organization Grant number Country National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) GM115386
Journal : J.Biol.Chem. / Year : 2016Title : Substrate-selective Inhibition of Cyclooxygeanse-2 by Fenamic Acid Derivatives Is Dependent on Peroxide Tone.Authors : Orlando, B.J. / Malkowski, M.G. History Deposition Mar 3, 2016 Deposition site / Processing site Revision 1.0 May 25, 2016 Provider / Type Revision 1.1 Jun 8, 2016 Group / Database referencesRevision 1.2 Jul 27, 2016 Group Revision 1.3 Sep 27, 2017 Group Advisory / Author supporting evidence ... Advisory / Author supporting evidence / Database references / Derived calculations Category citation / pdbx_audit_support ... citation / pdbx_audit_support / pdbx_struct_oper_list / pdbx_unobs_or_zero_occ_atoms Item / _pdbx_audit_support.funding_organization / _pdbx_struct_oper_list.symmetry_operationRevision 1.4 Dec 25, 2019 Group / Category / Item Revision 2.0 Jul 29, 2020 Group Advisory / Atomic model ... Advisory / Atomic model / Data collection / Derived calculations / Structure summary Category atom_site / chem_comp ... atom_site / chem_comp / entity / pdbx_branch_scheme / pdbx_chem_comp_identifier / pdbx_entity_branch / pdbx_entity_branch_descriptor / pdbx_entity_branch_link / pdbx_entity_branch_list / pdbx_entity_nonpoly / pdbx_nonpoly_scheme / pdbx_struct_assembly_gen / pdbx_struct_conn_angle / pdbx_struct_special_symmetry / pdbx_validate_close_contact / struct_asym / struct_conn / struct_site / struct_site_gen Item _atom_site.B_iso_or_equiv / _atom_site.Cartn_x ... _atom_site.B_iso_or_equiv / _atom_site.Cartn_x / _atom_site.Cartn_y / _atom_site.Cartn_z / _atom_site.auth_asym_id / _atom_site.auth_atom_id / _atom_site.auth_comp_id / _atom_site.auth_seq_id / _atom_site.label_asym_id / _atom_site.label_atom_id / _atom_site.label_comp_id / _atom_site.label_entity_id / _atom_site.type_symbol / _chem_comp.mon_nstd_flag / _chem_comp.name / _chem_comp.type / _entity.formula_weight / _entity.pdbx_description / _entity.pdbx_number_of_molecules / _entity.type / _pdbx_struct_assembly_gen.asym_id_list / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr2_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_special_symmetry.label_asym_id / _pdbx_validate_close_contact.auth_asym_id_2 / _pdbx_validate_close_contact.auth_seq_id_2 / _struct_conn.conn_type_id / _struct_conn.id / _struct_conn.pdbx_dist_value / _struct_conn.pdbx_leaving_atom_flag / _struct_conn.pdbx_role / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id Description / Provider / Type
Show all Show less
 Open data
Open data Basic information
Basic information Components
Components Cyclooxygenase
Cyclooxygenase  Keywords
Keywords OXIDOREDUCTASE / COX Mefenamic
OXIDOREDUCTASE / COX Mefenamic Function and homology information
Function and homology information hair cycle / positive regulation of fibroblast growth factor production /
hair cycle / positive regulation of fibroblast growth factor production /  prostaglandin-endoperoxide synthase ...Biosynthesis of EPA-derived SPMs / Biosynthesis of DPAn-3 SPMs / Biosynthesis of DHA-derived SPMs / Biosynthesis of electrophilic ω-3 PUFA oxo-derivatives / Synthesis of 15-eicosatetraenoic acid derivatives / cellular response to non-ionic osmotic stress / positive regulation of platelet-derived growth factor production /
prostaglandin-endoperoxide synthase ...Biosynthesis of EPA-derived SPMs / Biosynthesis of DPAn-3 SPMs / Biosynthesis of DHA-derived SPMs / Biosynthesis of electrophilic ω-3 PUFA oxo-derivatives / Synthesis of 15-eicosatetraenoic acid derivatives / cellular response to non-ionic osmotic stress / positive regulation of platelet-derived growth factor production /  hair cycle / positive regulation of fibroblast growth factor production /
hair cycle / positive regulation of fibroblast growth factor production /  prostaglandin-endoperoxide synthase /
prostaglandin-endoperoxide synthase /  prostaglandin-endoperoxide synthase activity / negative regulation of synaptic transmission, dopaminergic / cellular response to lead ion / response to nematode / positive regulation of transforming growth factor beta production / negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress / positive regulation of prostaglandin biosynthetic process / regulation of neuroinflammatory response / positive regulation of synaptic plasticity / Synthesis of Prostaglandins (PG) and Thromboxanes (TX) / oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen / response to fructose / positive regulation of smooth muscle contraction / Nicotinamide salvaging / response to fatty acid / cyclooxygenase pathway / positive regulation of fever generation / response to vitamin D / prostaglandin secretion / cellular response to fluid shear stress / nuclear outer membrane / response to angiotensin / response to manganese ion / prostaglandin biosynthetic process / nuclear inner membrane / cellular response to ATP / negative regulation of smooth muscle contraction / maintenance of blood-brain barrier / positive regulation of cell migration involved in sprouting angiogenesis /
prostaglandin-endoperoxide synthase activity / negative regulation of synaptic transmission, dopaminergic / cellular response to lead ion / response to nematode / positive regulation of transforming growth factor beta production / negative regulation of intrinsic apoptotic signaling pathway in response to osmotic stress / positive regulation of prostaglandin biosynthetic process / regulation of neuroinflammatory response / positive regulation of synaptic plasticity / Synthesis of Prostaglandins (PG) and Thromboxanes (TX) / oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen / response to fructose / positive regulation of smooth muscle contraction / Nicotinamide salvaging / response to fatty acid / cyclooxygenase pathway / positive regulation of fever generation / response to vitamin D / prostaglandin secretion / cellular response to fluid shear stress / nuclear outer membrane / response to angiotensin / response to manganese ion / prostaglandin biosynthetic process / nuclear inner membrane / cellular response to ATP / negative regulation of smooth muscle contraction / maintenance of blood-brain barrier / positive regulation of cell migration involved in sprouting angiogenesis /  bone mineralization / Interleukin-10 signaling / negative regulation of calcium ion transport /
bone mineralization / Interleukin-10 signaling / negative regulation of calcium ion transport /  decidualization / negative regulation of cell cycle / positive regulation of vascular endothelial growth factor production / response to tumor necrosis factor / brown fat cell differentiation / positive regulation of vasoconstriction / response to glucocorticoid /
decidualization / negative regulation of cell cycle / positive regulation of vascular endothelial growth factor production / response to tumor necrosis factor / brown fat cell differentiation / positive regulation of vasoconstriction / response to glucocorticoid /  embryo implantation / positive regulation of brown fat cell differentiation /
embryo implantation / positive regulation of brown fat cell differentiation /  peroxidase activity / positive regulation of synaptic transmission, glutamatergic /
peroxidase activity / positive regulation of synaptic transmission, glutamatergic /  learning /
learning /  caveola / positive regulation of smooth muscle cell proliferation /
caveola / positive regulation of smooth muscle cell proliferation /  memory /
memory /  regulation of blood pressure / positive regulation of protein import into nucleus / cellular response to mechanical stimulus / positive regulation of nitric oxide biosynthetic process / response to estradiol / cellular response to heat / positive regulation of peptidyl-serine phosphorylation /
regulation of blood pressure / positive regulation of protein import into nucleus / cellular response to mechanical stimulus / positive regulation of nitric oxide biosynthetic process / response to estradiol / cellular response to heat / positive regulation of peptidyl-serine phosphorylation /  regulation of inflammatory response / cellular response to hypoxia /
regulation of inflammatory response / cellular response to hypoxia /  angiogenesis / Interleukin-4 and Interleukin-13 signaling / response to oxidative stress / response to lipopolysaccharide / neuron projection / response to xenobiotic stimulus / positive regulation of apoptotic process /
angiogenesis / Interleukin-4 and Interleukin-13 signaling / response to oxidative stress / response to lipopolysaccharide / neuron projection / response to xenobiotic stimulus / positive regulation of apoptotic process /  endoplasmic reticulum lumen / negative regulation of cell population proliferation /
endoplasmic reticulum lumen / negative regulation of cell population proliferation /  heme binding / endoplasmic reticulum membrane /
heme binding / endoplasmic reticulum membrane /  enzyme binding /
enzyme binding /  endoplasmic reticulum / protein homodimerization activity / protein-containing complex /
endoplasmic reticulum / protein homodimerization activity / protein-containing complex /  metal ion binding /
metal ion binding /  cytoplasm
cytoplasm
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.342 Å
SYNCHROTRON / Resolution: 2.342 Å  Authors
Authors United States, 1items
United States, 1items  Citation
Citation Journal: J.Biol.Chem. / Year: 2016
Journal: J.Biol.Chem. / Year: 2016 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 5ikr.cif.gz
5ikr.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb5ikr.ent.gz
pdb5ikr.ent.gz PDB format
PDB format 5ikr.json.gz
5ikr.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/ik/5ikr
https://data.pdbj.org/pub/pdb/validation_reports/ik/5ikr ftp://data.pdbj.org/pub/pdb/validation_reports/ik/5ikr
ftp://data.pdbj.org/pub/pdb/validation_reports/ik/5ikr Links
Links Assembly
Assembly
 Components
Components Cyclooxygenase / Cyclooxygenase-2 / COX-2 / PHS II / Prostaglandin H2 synthase 2 / PGHS-2 / Prostaglandin- ...Cyclooxygenase-2 / COX-2 / PHS II / Prostaglandin H2 synthase 2 / PGHS-2 / Prostaglandin-endoperoxide synthase 2
Cyclooxygenase / Cyclooxygenase-2 / COX-2 / PHS II / Prostaglandin H2 synthase 2 / PGHS-2 / Prostaglandin- ...Cyclooxygenase-2 / COX-2 / PHS II / Prostaglandin H2 synthase 2 / PGHS-2 / Prostaglandin-endoperoxide synthase 2
 Homo sapiens (human) / Gene: PTGS2, COX2 / Production host:
Homo sapiens (human) / Gene: PTGS2, COX2 / Production host: 
 Spodoptera frugiperda (fall armyworm)
Spodoptera frugiperda (fall armyworm) prostaglandin-endoperoxide synthase
prostaglandin-endoperoxide synthase


 / Mass: 586.542 Da / Num. of mol.: 2
/ Mass: 586.542 Da / Num. of mol.: 2 N-Acetylglucosamine
N-Acetylglucosamine Octyl glucoside
Octyl glucoside





 Mefenamic acid
Mefenamic acid Ammonium
Ammonium Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  APS
APS  / Beamline: 17-ID / Wavelength: 1 Å
/ Beamline: 17-ID / Wavelength: 1 Å : 1 Å / Relative weight: 1
: 1 Å / Relative weight: 1  Processing
Processing Movie
Movie Controller
Controller

























 PDBj
PDBj











