[English] 日本語
 Yorodumi
Yorodumi- PDB-5ahi: Crystal structure of salmonalla enterica HisA mutant D7N with ProFAR -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 5ahi | ||||||
|---|---|---|---|---|---|---|---|
| Title | Crystal structure of salmonalla enterica HisA mutant D7N with ProFAR | ||||||
 Components Components | 1-(5-PHOSPHORIBOSYL)-5-[(5-PHOSPHORIBOSYLAMINO) METHYLIDENE AMINO] IMIDAZOLE-4-CARBOXAMIDE ISOMERASE | ||||||
 Keywords Keywords |  ISOMERASE / HISA / HISTIDINE BIOSYNTHESIS ISOMERASE / HISA / HISTIDINE BIOSYNTHESIS | ||||||
| Function / homology |  Function and homology information Function and homology information1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino]imidazole-4-carboxamide isomerase / 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino]imidazole-4-carboxamide isomerase activity / histidine biosynthetic process / tryptophan biosynthetic process /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   SALMONELLA ENTERICA (bacteria) SALMONELLA ENTERICA (bacteria) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.999 Å MOLECULAR REPLACEMENT / Resolution: 1.999 Å | ||||||
 Authors Authors | Soderholm, A. / Guo, X. / Newton, M.S. / Evans, G.B. / Nasvall, J. / Patrick, W.M. / Selmer, M. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: Structure and Mechanism of Hisa from Salmonella Enterica Authors: Soderholm, A. / Guo, X. / Newton, M.S. / Evans, G.B. / Nasvall, J. / Patrick, W.M. / Selmer, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  5ahi.cif.gz 5ahi.cif.gz | 100.8 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb5ahi.ent.gz pdb5ahi.ent.gz | 83.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  5ahi.json.gz 5ahi.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ah/5ahi https://data.pdbj.org/pub/pdb/validation_reports/ah/5ahi ftp://data.pdbj.org/pub/pdb/validation_reports/ah/5ahi ftp://data.pdbj.org/pub/pdb/validation_reports/ah/5ahi | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Unit cell |
| |||||||||
| Components on special symmetry positions |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein | Mass: 27130.943 Da / Num. of mol.: 1 / Mutation: YES Source method: isolated from a genetically manipulated source Source: (gene. exp.)   SALMONELLA ENTERICA (bacteria) / Plasmid: PEXP5-CT / Production host: SALMONELLA ENTERICA (bacteria) / Plasmid: PEXP5-CT / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): BL21 ESCHERICHIA COLI (E. coli) / Strain (production host): BL21References: UniProt: P10372, 1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)methylideneamino]imidazole-4-carboxamide isomerase |
|---|
-Non-polymers , 5 types, 110 molecules 
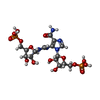







| #2: Chemical | ChemComp-GOL /  Glycerol Glycerol |
|---|---|
| #3: Chemical | ChemComp-GUO / [( |
| #4: Chemical | ChemComp-PO4 /  Phosphate Phosphate |
| #5: Chemical | ChemComp-CL /  Chloride Chloride |
| #6: Water | ChemComp-HOH /  Water Water |
-Details
| Nonpolymer details | GUO: PARTLY OBSERVED LIGAND PROFAR |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.38 Å3/Da / Density % sol: 48.39 % / Description: NONE |
|---|---|
Crystal grow | pH: 7 Details: 0.2M LITHIUM SULFATE, 30% W/V PEG 8K, 0.1M SODIUM ACETATE PH4.5, PH 7 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID23-2 / Wavelength: 0.8726 / Beamline: ID23-2 / Wavelength: 0.8726 |
| Detector | Type: DECTRIS PILATUS 6M / Detector: PIXEL / Date: Apr 27, 2014 / Details: PT COATED MIRRORS |
| Radiation | Monochromator: SI (111) / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.8726 Å / Relative weight: 1 : 0.8726 Å / Relative weight: 1 |
| Reflection | Resolution: 2→46.94 Å / Num. obs: 17983 / % possible obs: 97.5 % / Observed criterion σ(I): 2.94 / Redundancy: 8.5 % / Biso Wilson estimate: 26.84 Å2 / Rmerge(I) obs: 0.14 / Net I/σ(I): 13.8 |
| Reflection shell | Resolution: 2→2.12 Å / Redundancy: 8.01 % / Rmerge(I) obs: 0.79 / Mean I/σ(I) obs: 2.94 / % possible all: 79.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: THE WILD TYPE S. ENTERICA HISA Resolution: 1.999→46.942 Å / SU ML: 0.24 / σ(F): 1.34 / Phase error: 24.02 / Stereochemistry target values: ML Details: RESIDUES 17-24, 174, 178-179 AND 245- 253 ARE DISORDERED
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.9 Å / VDW probe radii: 1.11 Å / Solvent model: FLAT BULK SOLVENT MODEL | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 34.57 Å2 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.999→46.942 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS params. | Method: refined / Refine-ID: X-RAY DIFFRACTION
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement TLS group |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj




