[English] 日本語
 Yorodumi
Yorodumi- PDB-4mt2: COMPARISON OF THE NMR SOLUTION STRUCTURE AND THE X-RAY CRYSTAL ST... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 4mt2 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | COMPARISON OF THE NMR SOLUTION STRUCTURE AND THE X-RAY CRYSTAL STRUCTURE OF RAT METALLOTHIONEIN-2 | |||||||||
 Components Components | METALLOTHIONEIN ISOFORM II | |||||||||
 Keywords Keywords |  METALLOTHIONEIN METALLOTHIONEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationMetallothioneins bind metals / negative regulation of growth / detoxification of copper ion / intracellular zinc ion homeostasis / cellular response to zinc ion / cellular response to copper ion / cellular response to cadmium ion / zinc ion binding /  metal ion binding / metal ion binding /  nucleus / nucleus /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Rattus rattus (black rat) Rattus rattus (black rat) | |||||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2 Å X-RAY DIFFRACTION / Resolution: 2 Å | |||||||||
 Authors Authors | Robbins, A.H. / Stout, C.D. | |||||||||
 Citation Citation |  Journal: Proc.Natl.Acad.Sci.USA / Year: 1992 Journal: Proc.Natl.Acad.Sci.USA / Year: 1992Title: Comparison of the NMR solution structure and the x-ray crystal structure of rat metallothionein-2. Authors: Braun, W. / Vasak, M. / Robbins, A.H. / Stout, C.D. / Wagner, G. / Kagi, J.H. / Wuthrich, K. #1:  Journal: J.Mol.Biol. / Year: 1991 Journal: J.Mol.Biol. / Year: 1991Title: Refined Crystal Structure of Cd, Zn Metallothionein at 2.0 Angstroms Resolution Authors: Robbins, A.H. / Mcree, D.E. / Collett, M.Williamson S.A. / Xoung, N.H. / Furey, W.F. / Wang, B.C. / Stout, C.D. #2:  Journal: Science / Year: 1986 Journal: Science / Year: 1986Title: Crystal Structure Ofcd,Zn Metallothionein Authors: Furey, W.F. / Robbins, A.H. / Clancy, L.L. / Winge, D.R. / Wang, B.-C. / Stout, C.D. #3:  Journal: J.Biol.Chem. / Year: 1983 Journal: J.Biol.Chem. / Year: 1983Title: Single Crystals of Cadmium, Zinc Metallothionein Authors: Melis, K.A. / Carter, D.C. / Stout, C.D. / Winge, D.R. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4mt2.cif.gz 4mt2.cif.gz | 21.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4mt2.ent.gz pdb4mt2.ent.gz | 16.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4mt2.json.gz 4mt2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mt/4mt2 https://data.pdbj.org/pub/pdb/validation_reports/mt/4mt2 ftp://data.pdbj.org/pub/pdb/validation_reports/mt/4mt2 ftp://data.pdbj.org/pub/pdb/validation_reports/mt/4mt2 | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 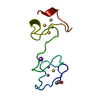
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein | Mass: 6179.380 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Rattus rattus (black rat) / References: UniProt: P04355 Rattus rattus (black rat) / References: UniProt: P04355 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| #2: Chemical | ChemComp-CD / #3: Chemical | #4: Chemical | ChemComp-NA / | #5: Water | ChemComp-HOH / |  Water WaterCompound details | TURN *T2* HAS A LEFT HANDED ALPHA CONFORMATI | Nonpolymer details | THE NUMBERING CONVENTION FOR THE FIVE CADMIUM ATOMS FOLLOWS THOSE USED IN THE NMR LITERATURE (SEE M. ...THE NUMBERING CONVENTION | |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.33 Å3/Da / Density % sol: 47.1 % | |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | *PLUS Temperature: 22 ℃ / pH: 7.5 / Method: vapor diffusion, hanging drop / Details: Melis, K.A., (1983) J.Biol.Chem., 258, 6255. | |||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Radiation | Scattering type: x-ray |
|---|---|
| Radiation wavelength | Relative weight: 1 |
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2→5 Å / Rfactor Rwork : 0.197 / σ(F): 0 : 0.197 / σ(F): 0 Details: THE STRUCTURE WAS SOLVED BY LOCATING THE FIVE CADMIUM ATOMS BY DIRECT METHODS. THE STARTING MODEL WAS A FIT OF THE CONSENSUS DOMAINS FROM THE 2-D NMR MODEL INTO AN ELECTRON DENSITY MAP ...Details: THE STRUCTURE WAS SOLVED BY LOCATING THE FIVE CADMIUM ATOMS BY DIRECT METHODS. THE STARTING MODEL WAS A FIT OF THE CONSENSUS DOMAINS FROM THE 2-D NMR MODEL INTO AN ELECTRON DENSITY MAP GENERATED FORM THE LOCATION OF THE CADMIUM POSITIONS. THE ANOMALOUS SCATTERING COMPONENTS OF CADMIUM ZINC AND SULFUR WERE INCLUDED IN THE REFINEMENT. DUE TO WEAK ELECTRON DENSITY, THE POSITIONS OF RESIDUES 1-2, 51-56, AND THE C-TERMINAL ALANINE SHOULD BE TREATED AS TENTATIVE. THE SIDE CHAINS OF LYSINES 20 AND 22 ARE ALSO UNRELIABLE. | ||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→5 Å
| ||||||||||||
| Refine LS restraints |
| ||||||||||||
| Software | *PLUS Name:  XPLOR / Classification: refinement XPLOR / Classification: refinement | ||||||||||||
| Refinement | *PLUS Rfactor Rwork : 0.197 : 0.197 | ||||||||||||
| Solvent computation | *PLUS | ||||||||||||
| Displacement parameters | *PLUS | ||||||||||||
| Refine LS restraints | *PLUS Type: x_angle_d / Dev ideal: 3.5 |
 Movie
Movie Controller
Controller










 PDBj
PDBj






