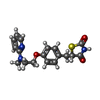
Entry Database : PDB / ID : 4emaTitle Human peroxisome proliferator-activated receptor gamma in complex with rosiglitazone Peroxisome proliferator-activated receptor gamma Keywords / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Homo sapiens (human)Method / / / Resolution : 2.545 Å Authors Liberato, M.V. / Nascimento, A.S. / Polikarpov, I. Journal : Plos One / Year : 2012Title : Medium chain fatty acids are selective peroxisome proliferator activated receptor (PPAR) Gamma activators and pan-PPAR partial agonistsAuthors: Liberato, M.V. / Nascimento, A.S. / Ayers, S.D. / Lin, J.Z. / Cvoro, A. / Silveira, R.L. / Martinez, L. / Souza, P.C. / Saidemberg, D. / Deng, T. / Amato, A.A. / Togashi, M. / Hsueh, W.A. / ... Authors : Liberato, M.V. / Nascimento, A.S. / Ayers, S.D. / Lin, J.Z. / Cvoro, A. / Silveira, R.L. / Martinez, L. / Souza, P.C. / Saidemberg, D. / Deng, T. / Amato, A.A. / Togashi, M. / Hsueh, W.A. / Phillips, K. / Palma, M.S. / Neves, F.A. / Skaf, M.S. / Webb, P. / Polikarpov, I. History Deposition Apr 11, 2012 Deposition site / Processing site Revision 1.0 Mar 6, 2013 Provider / Type Revision 1.1 Feb 28, 2024 Group / Database references / Derived calculationsCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / struct_ref_seq_dif / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_ref_seq_dif.details / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components
 Keywords
Keywords TRANSCRIPTION /
TRANSCRIPTION /  Nuclear receptor /
Nuclear receptor /  Retinoic acid receptor /
Retinoic acid receptor /  Nucleus
Nucleus Function and homology information
Function and homology information prostaglandin receptor activity / regulation of cholesterol transporter activity / negative regulation of connective tissue replacement involved in inflammatory response wound healing / negative regulation of receptor signaling pathway via STAT / MECP2 regulates transcription factors / negative regulation of extracellular matrix assembly / negative regulation of vascular endothelial cell proliferation / negative regulation of cellular response to transforming growth factor beta stimulus / negative regulation of cardiac muscle hypertrophy in response to stress /
prostaglandin receptor activity / regulation of cholesterol transporter activity / negative regulation of connective tissue replacement involved in inflammatory response wound healing / negative regulation of receptor signaling pathway via STAT / MECP2 regulates transcription factors / negative regulation of extracellular matrix assembly / negative regulation of vascular endothelial cell proliferation / negative regulation of cellular response to transforming growth factor beta stimulus / negative regulation of cardiac muscle hypertrophy in response to stress /  arachidonic acid binding ...
arachidonic acid binding ... prostaglandin receptor activity / regulation of cholesterol transporter activity / negative regulation of connective tissue replacement involved in inflammatory response wound healing / negative regulation of receptor signaling pathway via STAT / MECP2 regulates transcription factors / negative regulation of extracellular matrix assembly / negative regulation of vascular endothelial cell proliferation / negative regulation of cellular response to transforming growth factor beta stimulus / negative regulation of cardiac muscle hypertrophy in response to stress /
prostaglandin receptor activity / regulation of cholesterol transporter activity / negative regulation of connective tissue replacement involved in inflammatory response wound healing / negative regulation of receptor signaling pathway via STAT / MECP2 regulates transcription factors / negative regulation of extracellular matrix assembly / negative regulation of vascular endothelial cell proliferation / negative regulation of cellular response to transforming growth factor beta stimulus / negative regulation of cardiac muscle hypertrophy in response to stress /  arachidonic acid binding / positive regulation of low-density lipoprotein receptor activity / positive regulation of adiponectin secretion / lipoprotein transport / negative regulation of sequestering of triglyceride / positive regulation of vascular associated smooth muscle cell apoptotic process / macrophage derived foam cell differentiation /
arachidonic acid binding / positive regulation of low-density lipoprotein receptor activity / positive regulation of adiponectin secretion / lipoprotein transport / negative regulation of sequestering of triglyceride / positive regulation of vascular associated smooth muscle cell apoptotic process / macrophage derived foam cell differentiation /  DNA binding domain binding / STAT family protein binding / positive regulation of fatty acid metabolic process / response to lipid / negative regulation of SMAD protein signal transduction / LBD domain binding / negative regulation of type II interferon-mediated signaling pathway / negative regulation of cholesterol storage /
DNA binding domain binding / STAT family protein binding / positive regulation of fatty acid metabolic process / response to lipid / negative regulation of SMAD protein signal transduction / LBD domain binding / negative regulation of type II interferon-mediated signaling pathway / negative regulation of cholesterol storage /  E-box binding / alpha-actinin binding / negative regulation of blood vessel endothelial cell migration / lipid homeostasis / negative regulation of vascular associated smooth muscle cell proliferation /
E-box binding / alpha-actinin binding / negative regulation of blood vessel endothelial cell migration / lipid homeostasis / negative regulation of vascular associated smooth muscle cell proliferation /  R-SMAD binding / monocyte differentiation / positive regulation of cholesterol efflux / negative regulation of macrophage derived foam cell differentiation / cellular response to low-density lipoprotein particle stimulus / negative regulation of lipid storage / negative regulation of BMP signaling pathway / white fat cell differentiation / negative regulation of mitochondrial fission / positive regulation of fat cell differentiation / negative regulation of osteoblast differentiation / long-chain fatty acid transport / retinoic acid receptor signaling pathway / positive regulation of DNA binding /
R-SMAD binding / monocyte differentiation / positive regulation of cholesterol efflux / negative regulation of macrophage derived foam cell differentiation / cellular response to low-density lipoprotein particle stimulus / negative regulation of lipid storage / negative regulation of BMP signaling pathway / white fat cell differentiation / negative regulation of mitochondrial fission / positive regulation of fat cell differentiation / negative regulation of osteoblast differentiation / long-chain fatty acid transport / retinoic acid receptor signaling pathway / positive regulation of DNA binding /  cell fate commitment / BMP signaling pathway / nuclear retinoid X receptor binding / negative regulation of signaling receptor activity / regulation of cellular response to insulin stimulus / cell maturation / positive regulation of adipose tissue development / epithelial cell differentiation / peroxisome proliferator activated receptor signaling pathway / hormone-mediated signaling pathway / negative regulation of angiogenesis / response to nutrient / negative regulation of MAP kinase activity / fatty acid metabolic process / negative regulation of miRNA transcription / placenta development / Regulation of PTEN gene transcription / negative regulation of smooth muscle cell proliferation /
cell fate commitment / BMP signaling pathway / nuclear retinoid X receptor binding / negative regulation of signaling receptor activity / regulation of cellular response to insulin stimulus / cell maturation / positive regulation of adipose tissue development / epithelial cell differentiation / peroxisome proliferator activated receptor signaling pathway / hormone-mediated signaling pathway / negative regulation of angiogenesis / response to nutrient / negative regulation of MAP kinase activity / fatty acid metabolic process / negative regulation of miRNA transcription / placenta development / Regulation of PTEN gene transcription / negative regulation of smooth muscle cell proliferation /  transcription coregulator binding /
transcription coregulator binding /  peptide binding / negative regulation of transforming growth factor beta receptor signaling pathway / SUMOylation of intracellular receptors / mRNA transcription by RNA polymerase II / lipid metabolic process /
peptide binding / negative regulation of transforming growth factor beta receptor signaling pathway / SUMOylation of intracellular receptors / mRNA transcription by RNA polymerase II / lipid metabolic process /  regulation of circadian rhythm / PPARA activates gene expression / negative regulation of inflammatory response / DNA-binding transcription repressor activity, RNA polymerase II-specific /
regulation of circadian rhythm / PPARA activates gene expression / negative regulation of inflammatory response / DNA-binding transcription repressor activity, RNA polymerase II-specific /  regulation of blood pressure / Transcriptional regulation of white adipocyte differentiation / positive regulation of miRNA transcription / Nuclear Receptor transcription pathway / RNA polymerase II transcription regulator complex / cellular response to insulin stimulus / activation of cysteine-type endopeptidase activity involved in apoptotic process / rhythmic process / : /
regulation of blood pressure / Transcriptional regulation of white adipocyte differentiation / positive regulation of miRNA transcription / Nuclear Receptor transcription pathway / RNA polymerase II transcription regulator complex / cellular response to insulin stimulus / activation of cysteine-type endopeptidase activity involved in apoptotic process / rhythmic process / : /  nuclear receptor activity / positive regulation of DNA-binding transcription factor activity /
nuclear receptor activity / positive regulation of DNA-binding transcription factor activity /  glucose homeostasis / cellular response to hypoxia /
glucose homeostasis / cellular response to hypoxia /  double-stranded DNA binding / DNA-binding transcription activator activity, RNA polymerase II-specific / DNA-binding transcription factor binding / sequence-specific DNA binding /
double-stranded DNA binding / DNA-binding transcription activator activity, RNA polymerase II-specific / DNA-binding transcription factor binding / sequence-specific DNA binding /  cell differentiation /
cell differentiation /  nucleic acid binding /
nucleic acid binding /  receptor complex / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of gene expression /
receptor complex / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific / RNA polymerase II cis-regulatory region sequence-specific DNA binding / DNA-binding transcription factor activity / negative regulation of gene expression /  innate immune response / intracellular membrane-bounded organelle / negative regulation of DNA-templated transcription /
innate immune response / intracellular membrane-bounded organelle / negative regulation of DNA-templated transcription /  chromatin binding
chromatin binding
 Homo sapiens (human)
Homo sapiens (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.545 Å
MOLECULAR REPLACEMENT / Resolution: 2.545 Å  Authors
Authors Citation
Citation Journal: Plos One / Year: 2012
Journal: Plos One / Year: 2012 Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 4ema.cif.gz
4ema.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb4ema.ent.gz
pdb4ema.ent.gz PDB format
PDB format 4ema.json.gz
4ema.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/em/4ema
https://data.pdbj.org/pub/pdb/validation_reports/em/4ema ftp://data.pdbj.org/pub/pdb/validation_reports/em/4ema
ftp://data.pdbj.org/pub/pdb/validation_reports/em/4ema Links
Links Assembly
Assembly



 Components
Components / PPAR-gamma / Nuclear receptor subfamily 1 group C member 3
/ PPAR-gamma / Nuclear receptor subfamily 1 group C member 3
 Homo sapiens (human) / Gene: PPARG, NR1C3 / Production host:
Homo sapiens (human) / Gene: PPARG, NR1C3 / Production host: 
 Escherichia coli (E. coli) / References: UniProt: P37231
Escherichia coli (E. coli) / References: UniProt: P37231 Rosiglitazone
Rosiglitazone Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  LNLS
LNLS  / Beamline: W01B-MX2 / Wavelength: 1.46 Å
/ Beamline: W01B-MX2 / Wavelength: 1.46 Å : 1.46 Å / Relative weight: 1
: 1.46 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT / Resolution: 2.545→28.89 Å / SU ML: 0.36 / σ(F): 1.34 / Phase error: 24.4 / Stereochemistry target values: ML
MOLECULAR REPLACEMENT / Resolution: 2.545→28.89 Å / SU ML: 0.36 / σ(F): 1.34 / Phase error: 24.4 / Stereochemistry target values: ML Movie
Movie Controller
Controller
























 PDBj
PDBj