+ Open data
Open data
- Basic information
Basic information
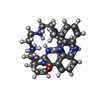
| Entry | Database: PDB / ID: 4bzv | ||||||
|---|---|---|---|---|---|---|---|
| Title | The Solution Structure of the MLN 944-d(TACGCGTA)2 complex | ||||||
 Components Components | DNA | ||||||
 Keywords Keywords |  DNA / BIS(PHENAZINE-1-CARBOXAMIDES) / MLN 944 / INTERCALATION / DNA / BIS(PHENAZINE-1-CARBOXAMIDES) / MLN 944 / INTERCALATION /  DRUG DESIGN / DRUG DESIGN /  ANTICANCER DRUG. ANTICANCER DRUG. | ||||||
| Function / homology | Chem-XR2 /  DNA DNA Function and homology information Function and homology information | ||||||
| Biological species | SYNTHETIC CONSTRUCT (others) | ||||||
| Method |  SOLUTION NMR / AMBER - MOLECULAR DYNAMICS SOLUTION NMR / AMBER - MOLECULAR DYNAMICS | ||||||
 Authors Authors | Serobian, A. / Thomas, D.S. / Ball, G.E. / Denny, W.A. / Wakelin, L.P.G. | ||||||
 Citation Citation |  Journal: Biopolymers / Year: 2014 Journal: Biopolymers / Year: 2014Title: The Solution Structure of Bis(Phenazine-1-Carboxamide)-DNA Complexes: Mln 944 Binding Corrected and Extended. Authors: Serobian, A. / Thomas, D.S. / Ball, G.E. / Denny, W.A. / Wakelin, L.P.G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  4bzv.cif.gz 4bzv.cif.gz | 18.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb4bzv.ent.gz pdb4bzv.ent.gz | 13.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  4bzv.json.gz 4bzv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/bz/4bzv https://data.pdbj.org/pub/pdb/validation_reports/bz/4bzv ftp://data.pdbj.org/pub/pdb/validation_reports/bz/4bzv ftp://data.pdbj.org/pub/pdb/validation_reports/bz/4bzv | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: DNA chain |  Mass: 2426.617 Da / Num. of mol.: 2 / Source method: obtained synthetically / Details: THE DNA CONFORMATION IS IN THE FORM OF B-DNA. / Source: (synth.) SYNTHETIC CONSTRUCT (others) #2: Chemical | ChemComp-XR2 / | |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||||||
| NMR details | Text: AVERAGE ENERGY MINIMISED SOLUTION STRUCTURE FROM 10 NS OF MOLECULAR DYNAMICS |
- Sample preparation
Sample preparation
| Details | Contents: 10% WATER/90% D2O |
|---|---|
| Sample conditions | Ionic strength: 80 mM / pH: 5.0 / Pressure: 1.0 atm / Temperature: 288.0 K |
-NMR measurement
| NMR spectrometer | Type: Bruker Avance / Manufacturer: Bruker / Model : Avance / Field strength: 700 MHz : Avance / Field strength: 700 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: AMBER - MOLECULAR DYNAMICS / Software ordinal: 1 | ||||||||||||
| NMR ensemble | Conformer selection criteria: DISTANCE RESTRAINTS / Conformers calculated total number: 1 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller






 PDBj
PDBj