[English] 日本語
 Yorodumi
Yorodumi- PDB-3zyb: CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALAG0 AT 2.3 A ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3zyb | ||||||
|---|---|---|---|---|---|---|---|
| Title | CRYSTAL STRUCTURE OF PA-IL LECTIN COMPLEXED WITH GALAG0 AT 2.3 A RESOLUTION | ||||||
 Components Components |
| ||||||
 Keywords Keywords | SUGAR BINDING PROTEIN / ADHESIN / GLYCOSPHINGOLIPID-ANTIGEN / GALACTOSE-SPECIFIC /  GALACTOSIDES GALACTOSIDES | ||||||
| Function / homology |  Function and homology information Function and homology informationheterophilic cell-cell adhesion via plasma membrane cell adhesion molecules /  carbohydrate binding / carbohydrate binding /  periplasmic space / periplasmic space /  cell surface / cell surface /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Pseudomonas aeruginosa (bacteria) Pseudomonas aeruginosa (bacteria)SYNTHETIC CONSTRUCT (others) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.29 Å MOLECULAR REPLACEMENT / Resolution: 2.29 Å | ||||||
 Authors Authors | Kadam, R.U. / Bergmann, M. / Hurley, M. / Garg, D. / Cacciarini, M. / Swiderska, M.A. / Nativi, C. / Sattler, M. / Smyth, A.R. / Williams, P. ...Kadam, R.U. / Bergmann, M. / Hurley, M. / Garg, D. / Cacciarini, M. / Swiderska, M.A. / Nativi, C. / Sattler, M. / Smyth, A.R. / Williams, P. / Camara, M. / Stocker, A. / Darbre, T. / Reymond, J.-L. | ||||||
 Citation Citation |  Journal: Angew.Chem.Int.Ed.Engl. / Year: 2011 Journal: Angew.Chem.Int.Ed.Engl. / Year: 2011Title: A Glycopeptide Dendrimer Inhibitor of the Galactose-Specific Lectin Leca and of Pseudomonas Aeruginosa Biofilms. Authors: Kadam, R.U. / Bergmann, M. / Hurley, M. / Garg, D. / Cacciarini, M. / Swiderska, M.A. / Nativi, C. / Sattler, M. / Smyth, A.R. / Williams, P. / Camara, M. / Stocker, A. / Darbre, T. / Reymond, J.-L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3zyb.cif.gz 3zyb.cif.gz | 217.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3zyb.ent.gz pdb3zyb.ent.gz | 171.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3zyb.json.gz 3zyb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/zy/3zyb https://data.pdbj.org/pub/pdb/validation_reports/zy/3zyb ftp://data.pdbj.org/pub/pdb/validation_reports/zy/3zyb ftp://data.pdbj.org/pub/pdb/validation_reports/zy/3zyb | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  3zyfC 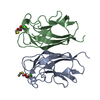 3zyhC  1l7lS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| 2 | 
| ||||||||
| 3 | 
| ||||||||
| 4 | 
| ||||||||
| 5 | 
| ||||||||
| 6 | 
| ||||||||
| 7 | 
| ||||||||
| 8 | 
| ||||||||
| Unit cell |
|
- Components
Components
-Protein / Protein/peptide / Sugars , 3 types, 19 molecules ABCDEFGHIJN

| #1: Protein | Mass: 12901.333 Da / Num. of mol.: 8 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria) Pseudomonas aeruginosa (strain ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1) (bacteria)Strain: ATCC 15692 / DSM 22644 / CIP 104116 / JCM 14847 / LMG 12228 / 1C / PRS 101 / PAO1 Gene: lecA, pa1L, PA2570 / Plasmid: PET25PAIL / Production host:   ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q05097 ESCHERICHIA COLI (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: Q05097#2: Protein/peptide | Mass: 355.475 Da / Num. of mol.: 3 / Source method: obtained synthetically / Details: GALA-LYS-PRO-LEUNH2 WAS SYNTHESIZED ON SOLID PHASE / Source: (synth.) SYNTHETIC CONSTRUCT (others) #4: Sugar | ChemComp-GAL /  Galactose Galactose |
|---|
-Non-polymers , 3 types, 959 molecules 
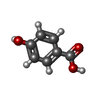



| #3: Chemical | ChemComp-CA / #5: Chemical | ChemComp-PHB /  4-Hydroxybenzoic acid 4-Hydroxybenzoic acid#6: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.4 Å3/Da / Density % sol: 48.9 % / Description: NONE |
|---|---|
Crystal grow | pH: 3.5 Details: 0.1 M CITRIC ACID PH 3.5 AND 25% W/V POLYETHYLENE GLYCOL 3350 |
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  SLS SLS  / Beamline: X06DA / Wavelength: 1 / Beamline: X06DA / Wavelength: 1 |
| Detector | Type: MARRESEARCH / Detector: CCD / Date: Aug 20, 2010 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1 Å / Relative weight: 1 : 1 Å / Relative weight: 1 |
| Reflection | Resolution: 2.29→73.07 Å / Num. obs: 45434 / % possible obs: 98.8 % / Observed criterion σ(I): 3 / Redundancy: 3.63 % / Biso Wilson estimate: 30 Å2 / Rmerge(I) obs: 0.1 / Net I/σ(I): 10 |
| Reflection shell | Resolution: 2.29→2.43 Å / Redundancy: 3.5 % / Rmerge(I) obs: 0.44 / Mean I/σ(I) obs: 2.8 / % possible all: 93.5 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1L7L Resolution: 2.29→49.261 Å / SU ML: 0.3 / σ(F): 1.99 / Phase error: 26.71 / Stereochemistry target values: ML
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Shrinkage radii: 0.61 Å / VDW probe radii: 0.9 Å / Solvent model: FLAT BULK SOLVENT MODEL / Bsol: 37.533 Å2 / ksol: 0.386 e/Å3 | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 26.2 Å2
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.29→49.261 Å
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj




