+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 3dar | ||||||
|---|---|---|---|---|---|---|---|
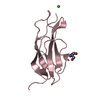
| Title | Crystal structure of D2 domain from human FGFR2 | ||||||
 Components Components | Fibroblast growth factor receptor 2 | ||||||
 Keywords Keywords |  TRANSFERASE / IMMUNOGLOBULIN FOLD / ATP-binding / TRANSFERASE / IMMUNOGLOBULIN FOLD / ATP-binding /  Craniosynostosis / Disease mutation / Craniosynostosis / Disease mutation /  Ectodermal dysplasia / Ectodermal dysplasia /  Glycoprotein / Heparin-binding / Glycoprotein / Heparin-binding /  Immunoglobulin domain / Immunoglobulin domain /  Kinase / Lacrimo-auriculo-dento-digital syndrome / Kinase / Lacrimo-auriculo-dento-digital syndrome /  Membrane / Nucleotide-binding / Membrane / Nucleotide-binding /  Phosphoprotein / Receptor / Phosphoprotein / Receptor /  Secreted / Secreted /  Transmembrane / Tyrosine-protein kinase Transmembrane / Tyrosine-protein kinase | ||||||
| Function / homology |  Function and homology information Function and homology informationSignaling by FGFR2 amplification mutants / Signaling by FGFR2 fusions / fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow cell / fibroblast growth factor receptor signaling pathway involved in hemopoiesis / fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow / fibroblast growth factor receptor signaling pathway involved in mammary gland specification / lateral sprouting from an epithelium / mammary gland bud formation / branch elongation involved in salivary gland morphogenesis / mesenchymal cell differentiation involved in lung development ...Signaling by FGFR2 amplification mutants / Signaling by FGFR2 fusions / fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow cell / fibroblast growth factor receptor signaling pathway involved in hemopoiesis / fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow / fibroblast growth factor receptor signaling pathway involved in mammary gland specification / lateral sprouting from an epithelium / mammary gland bud formation / branch elongation involved in salivary gland morphogenesis / mesenchymal cell differentiation involved in lung development / lacrimal gland development / otic vesicle formation / prostate gland morphogenesis / regulation of smooth muscle cell differentiation / orbitofrontal cortex development / regulation of morphogenesis of a branching structure / squamous basal epithelial stem cell differentiation involved in prostate gland acinus development / branching morphogenesis of a nerve / embryonic organ morphogenesis / endochondral bone growth / morphogenesis of embryonic epithelium / epidermis morphogenesis / bud elongation involved in lung branching / positive regulation of epithelial cell proliferation involved in lung morphogenesis / pyramidal neuron development / membranous septum morphogenesis / reproductive structure development / limb bud formation / lung lobe morphogenesis / gland morphogenesis / fibroblast growth factor receptor signaling pathway involved in orbitofrontal cortex development / ventricular zone neuroblast division / embryonic digestive tract morphogenesis / epithelial cell proliferation involved in salivary gland morphogenesis / mesenchymal cell differentiation / branching involved in prostate gland morphogenesis / mesenchymal cell proliferation involved in lung development / branching involved in labyrinthine layer morphogenesis / FGFR2b ligand binding and activation / FGFR2c ligand binding and activation / Activated point mutants of FGFR2 / regulation of osteoblast proliferation / Phospholipase C-mediated cascade; FGFR2 /  fibroblast growth factor receptor activity / branching involved in salivary gland morphogenesis / embryonic pattern specification / outflow tract septum morphogenesis / positive regulation of phospholipase activity / lung-associated mesenchyme development / mesodermal cell differentiation / digestive tract development / regulation of smoothened signaling pathway / embryonic cranial skeleton morphogenesis / bone morphogenesis / fibroblast growth factor receptor activity / branching involved in salivary gland morphogenesis / embryonic pattern specification / outflow tract septum morphogenesis / positive regulation of phospholipase activity / lung-associated mesenchyme development / mesodermal cell differentiation / digestive tract development / regulation of smoothened signaling pathway / embryonic cranial skeleton morphogenesis / bone morphogenesis /  skeletal system morphogenesis / skeletal system morphogenesis /  odontogenesis / ureteric bud development / positive regulation of mesenchymal cell proliferation / regulation of osteoblast differentiation / ventricular cardiac muscle tissue morphogenesis / inner ear morphogenesis / Signaling by FGFR2 IIIa TM / organ growth / midbrain development / hair follicle morphogenesis / lung alveolus development / odontogenesis / ureteric bud development / positive regulation of mesenchymal cell proliferation / regulation of osteoblast differentiation / ventricular cardiac muscle tissue morphogenesis / inner ear morphogenesis / Signaling by FGFR2 IIIa TM / organ growth / midbrain development / hair follicle morphogenesis / lung alveolus development /  fibroblast growth factor binding / prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis / PI-3K cascade:FGFR2 / fibroblast growth factor binding / prostate epithelial cord arborization involved in prostate glandular acinus morphogenesis / PI-3K cascade:FGFR2 /  bone mineralization / prostate epithelial cord elongation / positive regulation of cell division / bone mineralization / prostate epithelial cord elongation / positive regulation of cell division /  excitatory synapse / positive regulation of Wnt signaling pathway / PI3K Cascade / excitatory synapse / positive regulation of Wnt signaling pathway / PI3K Cascade /  epithelial to mesenchymal transition / negative regulation of keratinocyte proliferation / fibroblast growth factor receptor signaling pathway / embryonic organ development / epithelial to mesenchymal transition / negative regulation of keratinocyte proliferation / fibroblast growth factor receptor signaling pathway / embryonic organ development /  cell fate commitment / positive regulation of cell cycle / SHC-mediated cascade:FGFR2 / cellular response to retinoic acid / FRS-mediated FGFR2 signaling / positive regulation of cardiac muscle cell proliferation / positive regulation of vascular associated smooth muscle cell proliferation / cellular response to transforming growth factor beta stimulus / Signaling by FGFR2 in disease / epithelial cell differentiation / cell fate commitment / positive regulation of cell cycle / SHC-mediated cascade:FGFR2 / cellular response to retinoic acid / FRS-mediated FGFR2 signaling / positive regulation of cardiac muscle cell proliferation / positive regulation of vascular associated smooth muscle cell proliferation / cellular response to transforming growth factor beta stimulus / Signaling by FGFR2 in disease / epithelial cell differentiation /  axonogenesis / post-embryonic development / regulation of ERK1 and ERK2 cascade / positive regulation of epithelial cell proliferation / Negative regulation of FGFR2 signaling / animal organ morphogenesis / lung development / axonogenesis / post-embryonic development / regulation of ERK1 and ERK2 cascade / positive regulation of epithelial cell proliferation / Negative regulation of FGFR2 signaling / animal organ morphogenesis / lung development /  bone development / bone development /  receptor protein-tyrosine kinase / peptidyl-tyrosine phosphorylation / positive regulation of canonical Wnt signaling pathway receptor protein-tyrosine kinase / peptidyl-tyrosine phosphorylation / positive regulation of canonical Wnt signaling pathwaySimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / MOLECULAR REPLACEMENT /  molecular replacement / Resolution: 2.2 Å molecular replacement / Resolution: 2.2 Å | ||||||
 Authors Authors | Brown, A. / Blundell, T.L. | ||||||
 Citation Citation |  Journal: To be Published Journal: To be PublishedTitle: Crystal structure of the FGFR2 D2 domain Authors: Brown, A. / Blundell, T.L. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  3dar.cif.gz 3dar.cif.gz | 56.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb3dar.ent.gz pdb3dar.ent.gz | 39.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  3dar.json.gz 3dar.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/da/3dar https://data.pdbj.org/pub/pdb/validation_reports/da/3dar ftp://data.pdbj.org/pub/pdb/validation_reports/da/3dar ftp://data.pdbj.org/pub/pdb/validation_reports/da/3dar | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1e0oS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||||||||||||||||||||||||
| 2 | 
| ||||||||||||||||||||||||||||||
| Unit cell |
| ||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments: Component-ID: 1 / Ens-ID: 1 / Beg auth comp-ID: LYS / Beg label comp-ID: LYS / End auth comp-ID: VAL / End label comp-ID: VAL / Auth seq-ID: 151 - 249 / Label seq-ID: 7 - 105
NCS oper:
|
- Components
Components
| #1: Protein |  / FGFR-2 / Keratinocyte growth factor receptor 2 / CD332 antigen / FGFR-2 / Keratinocyte growth factor receptor 2 / CD332 antigenMass: 12183.853 Da / Num. of mol.: 2 Fragment: D2 domain, Ig-like C2-type 2, UNP residues 146-249 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: FGFR2, BEK, KGFR, KSAM / Plasmid: pBAT4 / Production host: Homo sapiens (human) / Gene: FGFR2, BEK, KGFR, KSAM / Plasmid: pBAT4 / Production host:   Escherichia coli (E. coli) / Strain (production host): Rosetta2 Escherichia coli (E. coli) / Strain (production host): Rosetta2References: UniProt: P21802,  receptor protein-tyrosine kinase receptor protein-tyrosine kinase#2: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.89 Å3/Da / Density % sol: 57.4 % |
|---|---|
Crystal grow | Temperature: 291 K / Method: vapor diffusion, hanging drop / pH: 8.5 Details: 0.2 M sodium acetate, 0.1 M Tris-HCl, 30% PEG 4000, VAPOR DIFFUSION, HANGING DROP, temperature 291.0K, pH 8.5 |
-Data collection
| Diffraction | Mean temperature: 100 K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  ESRF ESRF  / Beamline: ID29 / Wavelength: 0.9763 Å / Beamline: ID29 / Wavelength: 0.9763 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Detector | Type: ADSC QUANTUM 315 / Detector: CCD / Date: May 6, 2006 / Details: Mirrors | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation | Monochromator: SI(311) MONOCHROMATOR / Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Radiation wavelength | Wavelength : 0.9763 Å / Relative weight: 1 : 0.9763 Å / Relative weight: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection | Resolution: 2.2→85.9 Å / Num. all: 14756 / Num. obs: 14667 / % possible obs: 98.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 2 / Redundancy: 7.3 % / Biso Wilson estimate: 44.3 Å2 / Rmerge(I) obs: 0.08 / Rsym value: 0.08 / Net I/σ(I): 6.7 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Reflection shell | Diffraction-ID: 1
|
-Phasing
Phasing | Method:  molecular replacement molecular replacement | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Phasing MR | Model details: Phaser MODE: MR_AUTO
|
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PBD Entry 1E0O, chain B, residues 149-249 Resolution: 2.2→33.917 Å / FOM work R set: 0.787 / Isotropic thermal model: isotropic / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / Stereochemistry target values: mlhl
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Bsol: 58.254 Å2 / ksol: 0.352 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso max: 78.28 Å2 / Biso mean: 38.91 Å2 / Biso min: 20.58 Å2
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.2→33.917 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints NCS | NCS model details: chain B & A / Rms dev position: 0.035 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj





