Entry Database : PDB / ID : 3bxkTitle Crystal structure of the P/Q-type calcium channel (CaV2.1) IQ domain and CA2+calmodulin complex Calmodulin Voltage-dependent P/Q-type calcium channel subunit alpha-1A peptide Keywords / / / / / / / / / / / / / / / / / / / / / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species Rattus norvegicus (Norway rat)Method / / Resolution : 2.55 Å Authors Mori, M.X. / Vander Kooi, C.W. / Leahy, D.J. / Yue, D.T. Journal : To be Published Title : Crystal structure of the P/Q-type calcium channel (CaV2.1) IQ domain and CA2+calmodulin complexAuthors : Mori, M.X. / Vander Kooi, C.W. / Leahy, D.J. / Yue, D.T. History Deposition Jan 14, 2008 Deposition site / Processing site Revision 1.0 Mar 25, 2008 Provider / Type Revision 1.1 Jul 13, 2011 Group / Version format complianceRevision 1.2 Feb 21, 2024 Group / Database references / Derived calculationsCategory chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / struct_conn / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Keywords
Keywords MEMBRANE PROTEIN /
MEMBRANE PROTEIN /  SIGNALING PROTEIN /
SIGNALING PROTEIN /  ION CHANNEL /
ION CHANNEL /  CALMODULIN /
CALMODULIN /  CALCIUM CHANNEL / IQ DOMAIN / FACILLITATION / INACTIVATION / CALCIUM-DEPENDENT /
CALCIUM CHANNEL / IQ DOMAIN / FACILLITATION / INACTIVATION / CALCIUM-DEPENDENT /  VOLTAGE-GATED / ROBETTA / SIMULATIONS /
VOLTAGE-GATED / ROBETTA / SIMULATIONS /  Acetylation /
Acetylation /  Methylation /
Methylation /  Phosphoprotein / Calcium transport /
Phosphoprotein / Calcium transport /  Glycoprotein /
Glycoprotein /  Ion transport /
Ion transport /  Ionic channel /
Ionic channel /  Membrane /
Membrane /  Transmembrane / Transport /
Transmembrane / Transport /  Voltage-gated channel
Voltage-gated channel Function and homology information
Function and homology information regulation of store-operated calcium channel activity / regulation of high voltage-gated calcium channel activity / Presynaptic depolarization and calcium channel opening / : / regulation of response to tumor cell / positive regulation of autophagic cell death / DAPK1-calmodulin complex / : / establishment of protein localization to mitochondrial membrane / type 3 metabotropic glutamate receptor binding ...
regulation of store-operated calcium channel activity / regulation of high voltage-gated calcium channel activity / Presynaptic depolarization and calcium channel opening / : / regulation of response to tumor cell / positive regulation of autophagic cell death / DAPK1-calmodulin complex / : / establishment of protein localization to mitochondrial membrane / type 3 metabotropic glutamate receptor binding ... regulation of store-operated calcium channel activity / regulation of high voltage-gated calcium channel activity / Presynaptic depolarization and calcium channel opening / : / regulation of response to tumor cell / positive regulation of autophagic cell death / DAPK1-calmodulin complex / : / establishment of protein localization to mitochondrial membrane / type 3 metabotropic glutamate receptor binding / high voltage-gated calcium channel activity / establishment of protein localization to membrane /
regulation of store-operated calcium channel activity / regulation of high voltage-gated calcium channel activity / Presynaptic depolarization and calcium channel opening / : / regulation of response to tumor cell / positive regulation of autophagic cell death / DAPK1-calmodulin complex / : / establishment of protein localization to mitochondrial membrane / type 3 metabotropic glutamate receptor binding / high voltage-gated calcium channel activity / establishment of protein localization to membrane /  syntaxin binding / regulation of synaptic vesicle endocytosis / negative regulation of high voltage-gated calcium channel activity / regulation of synaptic vesicle exocytosis / negative regulation of calcium ion export across plasma membrane / organelle localization by membrane tethering / regulation of cardiac muscle cell action potential / mitochondrion-endoplasmic reticulum membrane tethering / autophagosome membrane docking / positive regulation of ryanodine-sensitive calcium-release channel activity /
syntaxin binding / regulation of synaptic vesicle endocytosis / negative regulation of high voltage-gated calcium channel activity / regulation of synaptic vesicle exocytosis / negative regulation of calcium ion export across plasma membrane / organelle localization by membrane tethering / regulation of cardiac muscle cell action potential / mitochondrion-endoplasmic reticulum membrane tethering / autophagosome membrane docking / positive regulation of ryanodine-sensitive calcium-release channel activity /  voltage-gated calcium channel complex /
voltage-gated calcium channel complex /  nitric-oxide synthase binding / protein phosphatase activator activity / calcium ion import across plasma membrane / calcium channel regulator activity / positive regulation of cyclic-nucleotide phosphodiesterase activity / response to amyloid-beta / positive regulation of phosphoprotein phosphatase activity /
nitric-oxide synthase binding / protein phosphatase activator activity / calcium ion import across plasma membrane / calcium channel regulator activity / positive regulation of cyclic-nucleotide phosphodiesterase activity / response to amyloid-beta / positive regulation of phosphoprotein phosphatase activity /  adenylate cyclase binding /
adenylate cyclase binding /  catalytic complex / detection of calcium ion / negative regulation of ryanodine-sensitive calcium-release channel activity /
catalytic complex / detection of calcium ion / negative regulation of ryanodine-sensitive calcium-release channel activity /  voltage-gated calcium channel activity / regulation of cardiac muscle contraction / calcium channel inhibitor activity / cellular response to interferon-beta / positive regulation of DNA binding / enzyme regulator activity /
voltage-gated calcium channel activity / regulation of cardiac muscle contraction / calcium channel inhibitor activity / cellular response to interferon-beta / positive regulation of DNA binding / enzyme regulator activity /  voltage-gated potassium channel complex /
voltage-gated potassium channel complex /  phosphatidylinositol 3-kinase binding / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / regulation of calcium-mediated signaling / positive regulation of protein dephosphorylation /
phosphatidylinositol 3-kinase binding / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / regulation of calcium-mediated signaling / positive regulation of protein dephosphorylation /  titin binding / regulation of ryanodine-sensitive calcium-release channel activity / potassium ion transmembrane transport / sperm midpiece / response to amphetamine /
titin binding / regulation of ryanodine-sensitive calcium-release channel activity / potassium ion transmembrane transport / sperm midpiece / response to amphetamine /  calcium channel complex / activation of adenylate cyclase activity / adenylate cyclase activator activity /
calcium channel complex / activation of adenylate cyclase activity / adenylate cyclase activator activity /  regulation of heart rate / nitric-oxide synthase regulator activity / protein serine/threonine kinase activator activity /
regulation of heart rate / nitric-oxide synthase regulator activity / protein serine/threonine kinase activator activity /  sarcomere /
sarcomere /  regulation of cytokinesis / cell projection / Regulation of insulin secretion / positive regulation of nitric-oxide synthase activity / calcium-mediated signaling / spindle microtubule / calcium ion transmembrane transport / positive regulation of receptor signaling pathway via JAK-STAT / modulation of chemical synaptic transmission /
regulation of cytokinesis / cell projection / Regulation of insulin secretion / positive regulation of nitric-oxide synthase activity / calcium-mediated signaling / spindle microtubule / calcium ion transmembrane transport / positive regulation of receptor signaling pathway via JAK-STAT / modulation of chemical synaptic transmission /  spindle pole / cellular response to type II interferon / response to calcium ion / calcium-dependent protein binding / cellular response to amyloid-beta / disordered domain specific binding / G2/M transition of mitotic cell cycle /
spindle pole / cellular response to type II interferon / response to calcium ion / calcium-dependent protein binding / cellular response to amyloid-beta / disordered domain specific binding / G2/M transition of mitotic cell cycle /  myelin sheath /
myelin sheath /  amyloid-beta binding / positive regulation of cytosolic calcium ion concentration /
amyloid-beta binding / positive regulation of cytosolic calcium ion concentration /  growth cone / chemical synaptic transmission / vesicle / transmembrane transporter binding / protein autophosphorylation / neuron projection / positive regulation of apoptotic process / protein domain specific binding /
growth cone / chemical synaptic transmission / vesicle / transmembrane transporter binding / protein autophosphorylation / neuron projection / positive regulation of apoptotic process / protein domain specific binding /  centrosome / neuronal cell body /
centrosome / neuronal cell body /  synapse /
synapse /  calcium ion binding /
calcium ion binding /  protein kinase binding / protein-containing complex /
protein kinase binding / protein-containing complex /  mitochondrion /
mitochondrion /  nucleoplasm /
nucleoplasm /  membrane /
membrane /  metal ion binding /
metal ion binding /  nucleus /
nucleus /  plasma membrane /
plasma membrane /  cytosol /
cytosol /  cytoplasm
cytoplasm
 Rattus norvegicus (Norway rat)
Rattus norvegicus (Norway rat) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.55 Å
MOLECULAR REPLACEMENT / Resolution: 2.55 Å  Authors
Authors Citation
Citation Journal: To be Published
Journal: To be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 3bxk.cif.gz
3bxk.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb3bxk.ent.gz
pdb3bxk.ent.gz PDB format
PDB format 3bxk.json.gz
3bxk.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/bx/3bxk
https://data.pdbj.org/pub/pdb/validation_reports/bx/3bxk ftp://data.pdbj.org/pub/pdb/validation_reports/bx/3bxk
ftp://data.pdbj.org/pub/pdb/validation_reports/bx/3bxk Links
Links Assembly
Assembly
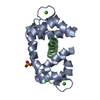

 Components
Components / CaM
/ CaM
 Rattus norvegicus (Norway rat) / Gene: Calm1, Calm, Cam, Cam1 / Plasmid: PET24B / Production host:
Rattus norvegicus (Norway rat) / Gene: Calm1, Calm, Cam, Cam1 / Plasmid: PET24B / Production host: 
 Escherichia coli (E. coli) / References: UniProt: P62161, UniProt: P0DP29*PLUS
Escherichia coli (E. coli) / References: UniProt: P62161, UniProt: P0DP29*PLUS Sulfate
Sulfate Water
Water X-RAY DIFFRACTION / Number of used crystals: 2
X-RAY DIFFRACTION / Number of used crystals: 2  Sample preparation
Sample preparation ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å
ROTATING ANODE / Type: RIGAKU RU200 / Wavelength: 1.5418 Å : 1.5418 Å / Relative weight: 1
: 1.5418 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT / Resolution: 2.55→24.87 Å / Cor.coef. Fo:Fc: 0.946 / Cor.coef. Fo:Fc free: 0.905 / SU B: 11.546 / SU ML: 0.251 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.655 / ESU R Free: 0.321 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS
MOLECULAR REPLACEMENT / Resolution: 2.55→24.87 Å / Cor.coef. Fo:Fc: 0.946 / Cor.coef. Fo:Fc free: 0.905 / SU B: 11.546 / SU ML: 0.251 / TLS residual ADP flag: LIKELY RESIDUAL / Cross valid method: THROUGHOUT / ESU R: 0.655 / ESU R Free: 0.321 / Stereochemistry target values: MAXIMUM LIKELIHOOD / Details: HYDROGENS HAVE BEEN ADDED IN THE RIDING POSITIONS Movie
Movie Controller
Controller







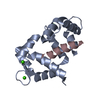





 PDBj
PDBj













