Entry Database : PDB / ID : 2xdgTitle Crystal structure of the extracellular domain of human growth hormone releasing hormone receptor. GROWTH HORMONE-RELEASING HORMONE RECEPTOR Keywords / / Function / homology Function Domain/homology Component
/ / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / / Biological species HOMO SAPIENS (human)Method / / / Resolution : 1.95 Å Authors Pike, A.C.W. / Quigley, A. / Barr, A.J. / Burgess Brown, N. / Shrestha, L. / Yang, J. / Chaikuad, A. / Vollmar, M. / Muniz, J.R.C. / von Delft, F. ...Pike, A.C.W. / Quigley, A. / Barr, A.J. / Burgess Brown, N. / Shrestha, L. / Yang, J. / Chaikuad, A. / Vollmar, M. / Muniz, J.R.C. / von Delft, F. / Edwards, A. / Arrowsmith, C.H. / Weigelt, J. / Bountra, C. / Carpenter, E.P. Journal : To be Published Title : Crystal Structure of the Extracellular Domain of Human Growth Hormone Releasing Hormone Receptor.Authors: Pike, A.C.W. / Quigley, A. / Barr, A.J. / Burgess Brown, N. / Shrestha, L. / Yang, J. / Chaikuad, A. / Vollmar, M. / Muniz, J.R.C. / von Delft, F. / Edwards, A. / Arrowsmith, C.H. / Weigelt, ... Authors : Pike, A.C.W. / Quigley, A. / Barr, A.J. / Burgess Brown, N. / Shrestha, L. / Yang, J. / Chaikuad, A. / Vollmar, M. / Muniz, J.R.C. / von Delft, F. / Edwards, A. / Arrowsmith, C.H. / Weigelt, J. / Bountra, C. / Carpenter, E.P. History Deposition Apr 30, 2010 Deposition site / Processing site Revision 1.0 Jun 16, 2010 Provider / Type Revision 1.1 May 8, 2011 Group Revision 1.2 Jul 13, 2011 Group Revision 1.3 Jan 24, 2018 Group / Structure summary / Category / citation_author / Item / _citation_author.nameRevision 1.4 Dec 20, 2023 Group Data collection / Database references ... Data collection / Database references / Derived calculations / Other / Refinement description Category chem_comp_atom / chem_comp_bond ... chem_comp_atom / chem_comp_bond / database_2 / pdbx_database_status / pdbx_initial_refinement_model / pdbx_struct_conn_angle / struct_conn / struct_site Item _database_2.pdbx_DOI / _database_2.pdbx_database_accession ... _database_2.pdbx_DOI / _database_2.pdbx_database_accession / _pdbx_database_status.status_code_sf / _pdbx_struct_conn_angle.ptnr1_auth_asym_id / _pdbx_struct_conn_angle.ptnr1_auth_comp_id / _pdbx_struct_conn_angle.ptnr1_auth_seq_id / _pdbx_struct_conn_angle.ptnr1_label_asym_id / _pdbx_struct_conn_angle.ptnr1_label_atom_id / _pdbx_struct_conn_angle.ptnr1_label_comp_id / _pdbx_struct_conn_angle.ptnr1_label_seq_id / _pdbx_struct_conn_angle.ptnr1_symmetry / _pdbx_struct_conn_angle.ptnr3_auth_asym_id / _pdbx_struct_conn_angle.ptnr3_auth_comp_id / _pdbx_struct_conn_angle.ptnr3_auth_seq_id / _pdbx_struct_conn_angle.ptnr3_label_asym_id / _pdbx_struct_conn_angle.ptnr3_label_atom_id / _pdbx_struct_conn_angle.ptnr3_label_comp_id / _pdbx_struct_conn_angle.ptnr3_label_seq_id / _pdbx_struct_conn_angle.ptnr3_symmetry / _pdbx_struct_conn_angle.value / _struct_conn.pdbx_dist_value / _struct_conn.ptnr1_auth_asym_id / _struct_conn.ptnr1_auth_comp_id / _struct_conn.ptnr1_auth_seq_id / _struct_conn.ptnr1_label_asym_id / _struct_conn.ptnr1_label_atom_id / _struct_conn.ptnr1_label_comp_id / _struct_conn.ptnr1_label_seq_id / _struct_conn.ptnr1_symmetry / _struct_conn.ptnr2_auth_asym_id / _struct_conn.ptnr2_auth_comp_id / _struct_conn.ptnr2_auth_seq_id / _struct_conn.ptnr2_label_asym_id / _struct_conn.ptnr2_label_atom_id / _struct_conn.ptnr2_label_comp_id / _struct_conn.ptnr2_label_seq_id / _struct_conn.ptnr2_symmetry / _struct_site.pdbx_auth_asym_id / _struct_site.pdbx_auth_comp_id / _struct_site.pdbx_auth_seq_id
Show all Show less
 Yorodumi
Yorodumi Open data
Open data Basic information
Basic information Components
Components Growth-hormone-releasing hormone receptor
Growth-hormone-releasing hormone receptor  Keywords
Keywords SIGNALING PROTEIN /
SIGNALING PROTEIN /  RECEPTOR /
RECEPTOR /  MEMBRANE PROTEIN
MEMBRANE PROTEIN Function and homology information
Function and homology information growth hormone-releasing hormone receptor activity / growth hormone secretion / positive regulation of growth hormone secretion / regulation of protein metabolic process / positive regulation of multicellular organism growth / multicellular organismal reproductive process / G protein-coupled peptide receptor activity / hormone metabolic process ...regulation of intracellular steroid hormone receptor signaling pathway / somatotropin secreting cell development /
growth hormone-releasing hormone receptor activity / growth hormone secretion / positive regulation of growth hormone secretion / regulation of protein metabolic process / positive regulation of multicellular organism growth / multicellular organismal reproductive process / G protein-coupled peptide receptor activity / hormone metabolic process ...regulation of intracellular steroid hormone receptor signaling pathway / somatotropin secreting cell development /  growth hormone-releasing hormone receptor activity / growth hormone secretion / positive regulation of growth hormone secretion / regulation of protein metabolic process / positive regulation of multicellular organism growth / multicellular organismal reproductive process / G protein-coupled peptide receptor activity / hormone metabolic process / nuclear outer membrane / positive regulation of insulin-like growth factor receptor signaling pathway / nuclear inner membrane /
growth hormone-releasing hormone receptor activity / growth hormone secretion / positive regulation of growth hormone secretion / regulation of protein metabolic process / positive regulation of multicellular organism growth / multicellular organismal reproductive process / G protein-coupled peptide receptor activity / hormone metabolic process / nuclear outer membrane / positive regulation of insulin-like growth factor receptor signaling pathway / nuclear inner membrane /  growth factor binding /
growth factor binding /  peptide hormone binding / cAMP-mediated signaling / cell maturation / response to glucocorticoid /
peptide hormone binding / cAMP-mediated signaling / cell maturation / response to glucocorticoid /  lactation /
lactation /  secretory granule / G protein-coupled receptor activity / establishment of localization in cell / determination of adult lifespan / response to insulin / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Glucagon-type ligand receptors /
secretory granule / G protein-coupled receptor activity / establishment of localization in cell / determination of adult lifespan / response to insulin / adenylate cyclase-activating G protein-coupled receptor signaling pathway / Glucagon-type ligand receptors /  nuclear matrix / response to estrogen / cellular response to insulin stimulus / G alpha (s) signalling events / cell surface receptor signaling pathway / positive regulation of cell population proliferation /
nuclear matrix / response to estrogen / cellular response to insulin stimulus / G alpha (s) signalling events / cell surface receptor signaling pathway / positive regulation of cell population proliferation /  cell surface /
cell surface /  membrane /
membrane /  plasma membrane /
plasma membrane /  cytoplasm
cytoplasm
 HOMO SAPIENS (human)
HOMO SAPIENS (human) X-RAY DIFFRACTION /
X-RAY DIFFRACTION /  SYNCHROTRON /
SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 1.95 Å
MOLECULAR REPLACEMENT / Resolution: 1.95 Å  Authors
Authors Citation
Citation Journal: To be Published
Journal: To be Published Structure visualization
Structure visualization Molmil
Molmil Jmol/JSmol
Jmol/JSmol Downloads & links
Downloads & links Download
Download 2xdg.cif.gz
2xdg.cif.gz PDBx/mmCIF format
PDBx/mmCIF format pdb2xdg.ent.gz
pdb2xdg.ent.gz PDB format
PDB format 2xdg.json.gz
2xdg.json.gz PDBx/mmJSON format
PDBx/mmJSON format Other downloads
Other downloads https://data.pdbj.org/pub/pdb/validation_reports/xd/2xdg
https://data.pdbj.org/pub/pdb/validation_reports/xd/2xdg ftp://data.pdbj.org/pub/pdb/validation_reports/xd/2xdg
ftp://data.pdbj.org/pub/pdb/validation_reports/xd/2xdg
 Links
Links Assembly
Assembly
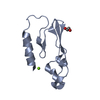

 Components
Components Growth-hormone-releasing hormone receptor / GHRH RECEPTOR / GROWTH HORMONE-RELEASING FACTOR GRF RECEPTOR / GRFR
Growth-hormone-releasing hormone receptor / GHRH RECEPTOR / GROWTH HORMONE-RELEASING FACTOR GRF RECEPTOR / GRFR
 HOMO SAPIENS (human) / Plasmid: PFB-SEC-NH / Cell line (production host): SF9 / Production host:
HOMO SAPIENS (human) / Plasmid: PFB-SEC-NH / Cell line (production host): SF9 / Production host: 
 SPODOPTERA FRUGIPERDA (fall armyworm) / References: UniProt: Q02643
SPODOPTERA FRUGIPERDA (fall armyworm) / References: UniProt: Q02643 Ethylene glycol
Ethylene glycol Water
Water X-RAY DIFFRACTION / Number of used crystals: 1
X-RAY DIFFRACTION / Number of used crystals: 1  Sample preparation
Sample preparation
 SYNCHROTRON / Site:
SYNCHROTRON / Site:  Diamond
Diamond  / Beamline: I24 / Wavelength: 0.9779
/ Beamline: I24 / Wavelength: 0.9779  : 0.9779 Å / Relative weight: 1
: 0.9779 Å / Relative weight: 1  Processing
Processing :
:  MOLECULAR REPLACEMENT
MOLECULAR REPLACEMENT Movie
Movie Controller
Controller










 PDBj
PDBj





