+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2mji | ||||||
|---|---|---|---|---|---|---|---|
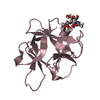
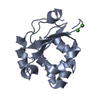
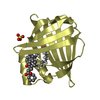
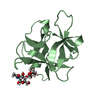
| Title | HIFABP_Ketorolac_complex | ||||||
 Components Components | Fatty acid-binding protein, intestinal | ||||||
 Keywords Keywords | LIPID BINDING PROTEIN /  TRANSPORT PROTEIN / TRANSPORT PROTEIN /  Protein-ligand complex / Intestinal Fatty Acid Binding Protein / Human FABP2 Protein-ligand complex / Intestinal Fatty Acid Binding Protein / Human FABP2 | ||||||
| Function / homology |  Function and homology information Function and homology informationintestinal lipid absorption / apical cortex / long-chain fatty acid transmembrane transporter activity /  long-chain fatty acid binding / Triglyceride catabolism / long-chain fatty acid binding / Triglyceride catabolism /  microvillus / fatty acid transport / fatty acid metabolic process / microvillus / fatty acid transport / fatty acid metabolic process /  fatty acid binding / fatty acid binding /  nucleus / nucleus /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing simulated annealing | ||||||
| Model details | closest to the average, model1 | ||||||
 Authors Authors | Patil, R. / Laguerre, A. / Wielens, J. / Headey, S. / Williams, M. / Mohanty, B. / Porter, C. / Scanlon, M. | ||||||
 Citation Citation |  Journal: Acs Chem.Biol. / Year: 2014 Journal: Acs Chem.Biol. / Year: 2014Title: Characterization of two distinct modes of drug binding to human intestinal Fatty Acid binding protein. Authors: Patil, R. / Laguerre, A. / Wielens, J. / Headey, S.J. / Williams, M.L. / Hughes, M.L. / Mohanty, B. / Porter, C.J. / Scanlon, M.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2mji.cif.gz 2mji.cif.gz | 423.1 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2mji.ent.gz pdb2mji.ent.gz | 362.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2mji.json.gz 2mji.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/mj/2mji https://data.pdbj.org/pub/pdb/validation_reports/mj/2mji ftp://data.pdbj.org/pub/pdb/validation_reports/mj/2mji ftp://data.pdbj.org/pub/pdb/validation_reports/mj/2mji | HTTPS FTP |
|---|
-Related structure data
| Similar structure data | |
|---|---|
| Other databases |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 15098.044 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: FABP2, FABPI / Production host: Homo sapiens (human) / Gene: FABP2, FABPI / Production host:   Escherichia coli (E. coli) / References: UniProt: P12104 Escherichia coli (E. coli) / References: UniProt: P12104 |
|---|---|
| #2: Chemical | ChemComp-KTR / ( Ketorolac Ketorolac |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||
| Sample conditions | pH: 5.5 / Pressure: ambient / Temperature: 298 K |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing / Software ordinal: 1 / Details: TORSIONAL ANGLE DYNAMICS simulated annealing / Software ordinal: 1 / Details: TORSIONAL ANGLE DYNAMICS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NMR constraints | NOE constraints total: 1591 / NOE intraresidue total count: 428 / NOE long range total count: 537 / NOE medium range total count: 186 / NOE sequential total count: 440 / Protein phi angle constraints total count: 84 / Protein psi angle constraints total count: 83 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NMR representative | Selection criteria: closest to the average | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: structures with the lowest energy Conformers calculated total number: 10 / Conformers submitted total number: 10 |
 Movie
Movie Controller
Controller









 PDBj
PDBj



