+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2jtn | ||||||
|---|---|---|---|---|---|---|---|
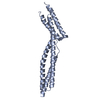
| Title | NMR Solution Structure of a ldb1-LID:Lhx3-LIM complex | ||||||
 Components Components | LIM domain-binding protein 1, LIM/homeobox protein Lhx3 | ||||||
 Keywords Keywords | PROTEIN BINDING/TRANSCRIPTION / intramolecular (fusion) protein-protein complex / PROTEIN BINDING-TRANSCRIPTION COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationmedial motor column neuron differentiation / prolactin secreting cell differentiation / somatotropin secreting cell differentiation / spinal cord motor neuron cell fate specification / ventral spinal cord interneuron specification /  regulation of kinase activity / spinal cord association neuron differentiation / cellular component assembly / negative regulation of erythrocyte differentiation / thyroid-stimulating hormone-secreting cell differentiation ...medial motor column neuron differentiation / prolactin secreting cell differentiation / somatotropin secreting cell differentiation / spinal cord motor neuron cell fate specification / ventral spinal cord interneuron specification / regulation of kinase activity / spinal cord association neuron differentiation / cellular component assembly / negative regulation of erythrocyte differentiation / thyroid-stimulating hormone-secreting cell differentiation ...medial motor column neuron differentiation / prolactin secreting cell differentiation / somatotropin secreting cell differentiation / spinal cord motor neuron cell fate specification / ventral spinal cord interneuron specification /  regulation of kinase activity / spinal cord association neuron differentiation / cellular component assembly / negative regulation of erythrocyte differentiation / thyroid-stimulating hormone-secreting cell differentiation / positive regulation of hemoglobin biosynthetic process / cerebellar Purkinje cell differentiation / head development / epithelial structure maintenance / primitive erythrocyte differentiation / transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery / beta-catenin-TCF complex / pituitary gland development / RUNX1 regulates transcription of genes involved in differentiation of HSCs / regulation of kinase activity / spinal cord association neuron differentiation / cellular component assembly / negative regulation of erythrocyte differentiation / thyroid-stimulating hormone-secreting cell differentiation / positive regulation of hemoglobin biosynthetic process / cerebellar Purkinje cell differentiation / head development / epithelial structure maintenance / primitive erythrocyte differentiation / transcription-dependent tethering of RNA polymerase II gene DNA at nuclear periphery / beta-catenin-TCF complex / pituitary gland development / RUNX1 regulates transcription of genes involved in differentiation of HSCs /  LIM domain binding / gastrulation with mouth forming second / dorsal/ventral pattern formation / motor neuron axon guidance / anterior/posterior axis specification / LIM domain binding / gastrulation with mouth forming second / dorsal/ventral pattern formation / motor neuron axon guidance / anterior/posterior axis specification /  regulation of focal adhesion assembly / cell leading edge / somatic stem cell population maintenance / inner ear development / positive regulation of cell adhesion / hair follicle development / regulation of focal adhesion assembly / cell leading edge / somatic stem cell population maintenance / inner ear development / positive regulation of cell adhesion / hair follicle development /  regulation of cell migration / cerebellum development / placenta development / regulation of cell migration / cerebellum development / placenta development /  transcription coregulator binding / positive regulation of transcription elongation by RNA polymerase II / RNA polymerase II transcription regulatory region sequence-specific DNA binding / lung development / transcription coregulator binding / positive regulation of transcription elongation by RNA polymerase II / RNA polymerase II transcription regulatory region sequence-specific DNA binding / lung development /  transcription coactivator binding / neuron differentiation / transcription coactivator binding / neuron differentiation /  Wnt signaling pathway / RNA polymerase II transcription regulator complex / : / Wnt signaling pathway / RNA polymerase II transcription regulator complex / : /  nervous system development / DNA-binding transcription activator activity, RNA polymerase II-specific / DNA-binding transcription factor binding / nervous system development / DNA-binding transcription activator activity, RNA polymerase II-specific / DNA-binding transcription factor binding /  transcription regulator complex / RNA polymerase II-specific DNA-binding transcription factor binding / sequence-specific DNA binding / transcription by RNA polymerase II / transcription regulator complex / RNA polymerase II-specific DNA-binding transcription factor binding / sequence-specific DNA binding / transcription by RNA polymerase II /  transcription coactivator activity / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific / transcription coactivator activity / transcription cis-regulatory region binding / DNA-binding transcription factor activity, RNA polymerase II-specific /  cell adhesion / negative regulation of DNA-templated transcription / apoptotic process / cell adhesion / negative regulation of DNA-templated transcription / apoptotic process /  chromatin binding / chromatin binding /  chromatin / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II / chromatin / regulation of transcription by RNA polymerase II / negative regulation of apoptotic process / positive regulation of DNA-templated transcription / negative regulation of transcription by RNA polymerase II /  enzyme binding / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / protein-containing complex / zinc ion binding / enzyme binding / protein homodimerization activity / positive regulation of transcription by RNA polymerase II / protein-containing complex / zinc ion binding /  nucleoplasm / identical protein binding / nucleoplasm / identical protein binding /  nucleus nucleusSimilarity search - Function | ||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing, torsion angle dynamics, cartesian dynamics simulated annealing, torsion angle dynamics, cartesian dynamics | ||||||
 Authors Authors | Lee, C. / Nancarrow, A.L. / Mackay, J.P. / Matthews, J.M. | ||||||
 Citation Citation |  Journal: Embo J. / Year: 2008 Journal: Embo J. / Year: 2008Title: Implementing the LIM code: the structural basis for cell type-specific assembly of LIM-homeodomain complexes Authors: Bhati, M. / Lee, C. / Nancarrow, A.L. / Lee, M. / Craig, V.J. / Bach, I. / Guss, J.M. / Mackay, J.P. / Matthews, J.M. | ||||||
| History |
| ||||||
| Remark 999 | SEQUENCE The sequence is composed of LIM domain-binding protein 1 and LIM/homeobox protein Lhx3 ... SEQUENCE The sequence is composed of LIM domain-binding protein 1 and LIM/homeobox protein Lhx3 with a linker, GGSGGHMGSGG. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2jtn.cif.gz 2jtn.cif.gz | 1 MB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2jtn.ent.gz pdb2jtn.ent.gz | 919.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2jtn.json.gz 2jtn.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/jt/2jtn https://data.pdbj.org/pub/pdb/validation_reports/jt/2jtn ftp://data.pdbj.org/pub/pdb/validation_reports/jt/2jtn ftp://data.pdbj.org/pub/pdb/validation_reports/jt/2jtn | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 20263.014 Da / Num. of mol.: 1 Fragment: Fusion of Ldb1 residues 295-340 and Lhx3 LIM zinc-binding domains 1 and 2 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Gene: Ldb1, Nli, Lhx3, Lim-3, Lim3, Plim / Production host: Mus musculus (house mouse) / Gene: Ldb1, Nli, Lhx3, Lim-3, Lim3, Plim / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: P70662, UniProt: P50481 Escherichia coli (E. coli) / Strain (production host): BL21(DE3) / References: UniProt: P70662, UniProt: P50481 |
|---|---|
| #2: Chemical | ChemComp-ZN / |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Details |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample |
|
 Movie
Movie Controller
Controller













 PDBj
PDBj



