[English] 日本語
 Yorodumi
Yorodumi- PDB-2fot: Crystal structure of the complex between calmodulin and alphaII-s... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 2fot | ||||||
|---|---|---|---|---|---|---|---|
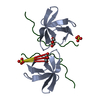
| Title | Crystal structure of the complex between calmodulin and alphaII-spectrin | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  METAL BINDING / METAL BINDING /  STRUCTURAL PROTEIN / STRUCTURAL PROTEIN /  calmodulin / calmodulin binding domain / calmodulin / calmodulin binding domain /  spectrin spectrin | ||||||
| Function / homology |  Function and homology information Function and homology information spectrin / actin filament capping / Sensory processing of sound by outer hair cells of the cochlea / Interaction between L1 and Ankyrins / Sensory processing of sound by inner hair cells of the cochlea / Nephrin family interactions / RHOV GTPase cycle / cortical actin cytoskeleton / positive regulation of ryanodine-sensitive calcium-release channel activity / RHOU GTPase cycle ... spectrin / actin filament capping / Sensory processing of sound by outer hair cells of the cochlea / Interaction between L1 and Ankyrins / Sensory processing of sound by inner hair cells of the cochlea / Nephrin family interactions / RHOV GTPase cycle / cortical actin cytoskeleton / positive regulation of ryanodine-sensitive calcium-release channel activity / RHOU GTPase cycle ... spectrin / actin filament capping / Sensory processing of sound by outer hair cells of the cochlea / Interaction between L1 and Ankyrins / Sensory processing of sound by inner hair cells of the cochlea / Nephrin family interactions / RHOV GTPase cycle / cortical actin cytoskeleton / positive regulation of ryanodine-sensitive calcium-release channel activity / RHOU GTPase cycle / negative regulation of ryanodine-sensitive calcium-release channel activity / Caspase-mediated cleavage of cytoskeletal proteins / COPI-mediated anterograde transport / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / NCAM signaling for neurite out-growth / cell projection / structural constituent of cytoskeleton / spectrin / actin filament capping / Sensory processing of sound by outer hair cells of the cochlea / Interaction between L1 and Ankyrins / Sensory processing of sound by inner hair cells of the cochlea / Nephrin family interactions / RHOV GTPase cycle / cortical actin cytoskeleton / positive regulation of ryanodine-sensitive calcium-release channel activity / RHOU GTPase cycle / negative regulation of ryanodine-sensitive calcium-release channel activity / Caspase-mediated cleavage of cytoskeletal proteins / COPI-mediated anterograde transport / regulation of release of sequestered calcium ion into cytosol by sarcoplasmic reticulum / NCAM signaling for neurite out-growth / cell projection / structural constituent of cytoskeleton /  spindle pole / specific granule lumen / spindle pole / specific granule lumen /  extracellular vesicle / microtubule cytoskeleton / extracellular vesicle / microtubule cytoskeleton /  actin filament binding / tertiary granule lumen / actin filament binding / tertiary granule lumen /  cell junction / cell junction /  actin binding / actin cytoskeleton organization / RAF/MAP kinase cascade / actin binding / actin cytoskeleton organization / RAF/MAP kinase cascade /  calmodulin binding / calmodulin binding /  cadherin binding / protein domain specific binding / intracellular membrane-bounded organelle / cadherin binding / protein domain specific binding / intracellular membrane-bounded organelle /  calcium ion binding / Neutrophil degranulation / protein-containing complex / extracellular exosome / extracellular region / calcium ion binding / Neutrophil degranulation / protein-containing complex / extracellular exosome / extracellular region /  membrane / membrane /  plasma membrane / plasma membrane /  cytosol / cytosol /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human)  Bos taurus (cattle) Bos taurus (cattle) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.45 Å MOLECULAR REPLACEMENT / Resolution: 2.45 Å | ||||||
 Authors Authors | Simonovic, M. / Zhang, Z. / Cianci, C.D. / Steitz, T.A. / Morrow, J.S. | ||||||
 Citation Citation |  Journal: J.Biol.Chem. / Year: 2006 Journal: J.Biol.Chem. / Year: 2006Title: Structure of the calmodulin alphaII-spectrin complex provides insight into the regulation of cell plasticity. Authors: Simonovic, M. / Zhang, Z. / Cianci, C.D. / Steitz, T.A. / Morrow, J.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  2fot.cif.gz 2fot.cif.gz | 47.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb2fot.ent.gz pdb2fot.ent.gz | 31.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  2fot.json.gz 2fot.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/fo/2fot https://data.pdbj.org/pub/pdb/validation_reports/fo/2fot ftp://data.pdbj.org/pub/pdb/validation_reports/fo/2fot ftp://data.pdbj.org/pub/pdb/validation_reports/fo/2fot | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1cdmS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Details | The biological unit is a monomer. |
- Components
Components
| #1: Protein |  / CaM / CaMMass: 16721.350 Da / Num. of mol.: 1 / Source method: isolated from a natural source / Source: (natural)   Bos taurus (cattle) / References: UniProt: P62157 Bos taurus (cattle) / References: UniProt: P62157 | ||
|---|---|---|---|
| #2: Protein/peptide | Mass: 4792.410 Da / Num. of mol.: 1 / Fragment: Calmodulin binding domain / Mutation: L1211R Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Gene: SPTAN1, SPTA2 / Plasmid: pGEX2T / Production host: Homo sapiens (human) / Gene: SPTAN1, SPTA2 / Plasmid: pGEX2T / Production host:   Escherichia coli (E. coli) / References: UniProt: Q13813 Escherichia coli (E. coli) / References: UniProt: Q13813 | ||
| #3: Chemical | ChemComp-CA / #4: Water | ChemComp-HOH / |  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.07 Å3/Da / Density % sol: 40.64 % |
|---|---|
Crystal grow | Temperature: 298 K / Method: vapor diffusion, sitting drop / pH: 8 Details: 50mM Tris,pH 8.00, 30% PEG 8000, 0.1M ammonium-sulfate, VAPOR DIFFUSION, SITTING DROP, temperature 298K |
-Data collection
| Diffraction |
| ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| ||||||||||||||||||
| Detector |
| ||||||||||||||||||
| Radiation |
| ||||||||||||||||||
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 | ||||||||||||||||||
| Reflection | Resolution: 2.45→45 Å / Num. all: 7933 / Num. obs: 7933 / % possible obs: 89.8 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 5.5 % / Biso Wilson estimate: 36.2 Å2 / Rmerge(I) obs: 0.245 / Net I/σ(I): 4.55 | ||||||||||||||||||
| Reflection shell | Resolution: 2.45→2.6 Å / Redundancy: 4.6 % / Rmerge(I) obs: 0.467 / Mean I/σ(I) obs: 2.2 / Num. unique all: 986 / % possible all: 87.1 |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: pdb entry 1cdm Resolution: 2.45→44.48 Å / Rfactor Rfree error: 0.01 / Data cutoff high absF: 551349.9 / Data cutoff low absF: 0 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / Stereochemistry target values: Engh & Huber
| |||||||||||||||||||||||||
| Solvent computation | Solvent model: FLAT MODEL / Bsol: 60.4525 Å2 / ksol: 0.355356 e/Å3 | |||||||||||||||||||||||||
| Displacement parameters | Biso mean: 44.3 Å2
| |||||||||||||||||||||||||
| Refine analyze |
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.45→44.48 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| LS refinement shell | Resolution: 2.45→2.6 Å / Rfactor Rfree error: 0.036 / Total num. of bins used: 6
| |||||||||||||||||||||||||
| Xplor file |
|
 Movie
Movie Controller
Controller














 PDBj
PDBj














