+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1tyr | ||||||
|---|---|---|---|---|---|---|---|
| Title | TRANSTHYRETIN COMPLEX WITH RETINOIC ACID | ||||||
 Components Components | TRANSTHYRETIN | ||||||
 Keywords Keywords | RETINOL-BINDING /  RETINOL-BINDING PROTEIN RETINOL-BINDING PROTEIN | ||||||
| Function / homology |  Function and homology information Function and homology informationRetinoid cycle disease events /  thyroid hormone binding / The canonical retinoid cycle in rods (twilight vision) / Non-integrin membrane-ECM interactions / purine nucleobase metabolic process / Retinoid metabolism and transport / thyroid hormone binding / The canonical retinoid cycle in rods (twilight vision) / Non-integrin membrane-ECM interactions / purine nucleobase metabolic process / Retinoid metabolism and transport /  hormone activity / azurophil granule lumen / Amyloid fiber formation / Neutrophil degranulation ...Retinoid cycle disease events / hormone activity / azurophil granule lumen / Amyloid fiber formation / Neutrophil degranulation ...Retinoid cycle disease events /  thyroid hormone binding / The canonical retinoid cycle in rods (twilight vision) / Non-integrin membrane-ECM interactions / purine nucleobase metabolic process / Retinoid metabolism and transport / thyroid hormone binding / The canonical retinoid cycle in rods (twilight vision) / Non-integrin membrane-ECM interactions / purine nucleobase metabolic process / Retinoid metabolism and transport /  hormone activity / azurophil granule lumen / Amyloid fiber formation / Neutrophil degranulation / hormone activity / azurophil granule lumen / Amyloid fiber formation / Neutrophil degranulation /  extracellular space / extracellular exosome / extracellular region / identical protein binding extracellular space / extracellular exosome / extracellular region / identical protein bindingSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 1.8 Å X-RAY DIFFRACTION / Resolution: 1.8 Å | ||||||
 Authors Authors | Zanotti, G. / D'Acunto, M.R. / Malpeli, G. / Folli, C. / Berni, R. | ||||||
 Citation Citation |  Journal: Eur.J.Biochem. / Year: 1995 Journal: Eur.J.Biochem. / Year: 1995Title: Crystal structure of the transthyretin--retinoic-acid complex Authors: Zanotti, G. / D'Acunto, M.R. / Malpeli, G. / Folli, C. / Berni, R. #1:  Journal: J.Biol.Chem. / Year: 1994 Journal: J.Biol.Chem. / Year: 1994Title: Crystallographic Studies on Complexes between Retinoids and Plasma Retinol-Binding Protein Authors: Zanotti, G. / Marcello, M. / Malpeli, G. / Folli, C. / Sartori, G. / Berni, R. #2:  Journal: Proc.Natl.Acad.Sci.USA / Year: 1992 Journal: Proc.Natl.Acad.Sci.USA / Year: 1992Title: Crystal Structure Determination at 2.3 Angstroms of Human Transthyretin-3',5'-Dibromo-2',4,4', 6-Tetra-Hydroxyaurone Complex Authors: Ciszak, E. / Cody, V. / Luft, J.R. #3:  Journal: J.Biol.Chem. / Year: 1992 Journal: J.Biol.Chem. / Year: 1992Title: Mechanism of Molecular Recognition. Structural Aspects of 3,3'-Diiodo-L-Thyronine Binding to Human Serum Transthyretin Authors: Wojtczak, A. / Luft, J. / Cody, V. #4:  Journal: Nature / Year: 1977 Journal: Nature / Year: 1977Title: Protein-DNA and Protein-Hormone Interactions in Prealbumin: A Model of the Thyroid Hormone Nuclear Receptor? Authors: Blake, C.C.F. / Oatley, S.J. #5:  Journal: J.Mol.Biol. / Year: 1974 Journal: J.Mol.Biol. / Year: 1974Title: Structure of Human Plasma Prealbumin at 2.5 Angstroms Resolution, a Preliminary Report on the Polypeptide Chain Conformation, Quaternary Structure and Thyroxine Binding Authors: Blake, C.C.F. / Geisow, M.J. / Swan, I.D.A. / Rerat, C. / Rerat, B. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1tyr.cif.gz 1tyr.cif.gz | 60.7 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1tyr.ent.gz pdb1tyr.ent.gz | 49.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1tyr.json.gz 1tyr.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ty/1tyr https://data.pdbj.org/pub/pdb/validation_reports/ty/1tyr ftp://data.pdbj.org/pub/pdb/validation_reports/ty/1tyr ftp://data.pdbj.org/pub/pdb/validation_reports/ty/1tyr | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| 2 | 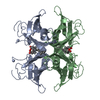
| ||||||||
| Unit cell |
| ||||||||
| Components on special symmetry positions |
| ||||||||
| Details | THE ENTIRE PROTEIN IS A TETRAMER, BUT THE ASYMMETRIC UNIT CONTAINS A DIMER. THE TWO DIMERS ARE RELATED BY A TWO FODL AXIS, WHILST THE TWO INDEPENDENT MONOMERS IN THE DIMER ARE RELATED BY A PSEUDO TWO-FOLD AXIS. THE LIGAND IS BOUND IN THE INTERNAL CHANNEL, MADE UP BY THE TWO INDEPENDENT MONOMERS. A CRYSTALLOGRAPHIC TWO-FOLD AXIS RUNS THROUGH THE CHANNEL, MAKING IT SYMMETRIC. MOREOVER, PERPENDICULAR TO THE PREVIOUS ONE, THERE IS THE PSEUDO TWO-FOLD, WHICH MAKES THE TWO HALFS OF THE CHANNEL NEARLY IDENTICAL. AS A RESULT OF ALL THAT, TWO INDEPENDENT BINDING SITES ARE PRESENT PER TERAMER, BUT, SINCE THE LIGAND IN THIS CASE DOES NOT PRESENT ANY SYMMETRY, EACH LIGAND IN THE CRYSTAL APPEARS TO BE SUPERIMPOSED TO ITS SYMMETRY-RELATED. IN CONCLUSION, FOUR RETINOIC ACID BOUND MOLECULES ARE SEEN, EACH PAIRS SUPERIMPOSED. IN ADDITION, SOLUTION DATA STRONGLY SUGGEST THAT ONLY ONE LIGAND MOLECULE IS BOUND TO A TTR TETRAMER. IN CONCLUSION, THE COORDINATES OF TWO 9CR MOLECULES ARE GIVEN, WHICH APPLYING THE CRYSTALLOGRAPHIC SYMMETRY, BECOMES FOUR, BUT ONLY ONE MUST BE CONSIDERED STATISTICALLY PRESENT IN THE TETRAMER. |
- Components
Components
| #1: Protein |  / PREALBUMIN / PREALBUMINMass: 13777.360 Da / Num. of mol.: 2 / Source method: isolated from a natural source / Source: (natural)   Homo sapiens (human) / References: UniProt: P02766 Homo sapiens (human) / References: UniProt: P02766#2: Chemical |  Alitretinoin Alitretinoin#3: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.25 Å3/Da / Density % sol: 45.39 % | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | *PLUS pH: 7 / Method: vapor diffusion, sitting drop | |||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction source | Wavelength: 1.5418 |
|---|---|
| Detector | Type: SIEMENS-NICOLET X100 / Detector: AREA DETECTOR / Date: Nov 1, 1994 |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Redundancy: 3 % / Rmerge(I) obs: 0.07 |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.8→9 Å / σ(F): 0 Details: THE ENTRY CONTAINS RETINOIC ACID MOLECULES IN BOTH BINDING SITES. THE OCCUPANCIES OF SITES WERE NOT REFINED, SINCE THE ELECTRON DENSITY REFLECTS THE PRESENCE OF THE CRYSTALLOGRAPHIC TWO-FOLD ...Details: THE ENTRY CONTAINS RETINOIC ACID MOLECULES IN BOTH BINDING SITES. THE OCCUPANCIES OF SITES WERE NOT REFINED, SINCE THE ELECTRON DENSITY REFLECTS THE PRESENCE OF THE CRYSTALLOGRAPHIC TWO-FOLD AXIS. IN THE FUNCTIONAL TETRAMER THE RETINOIC ACID MOLECULES ARE DISORDERED DUE TO THE CRYSTALLOGRAPHIC TWO-FOLD RUNNING THROUGH THE BINDING SITE. RESIDUES 1 - 9 OF BOTH CHAINS, AS WELL AS 126 - 127 OF CHAIN B, ARE ILL-DEFINED IN THE ELECTRON DENSITY. THEY HAVE BEEN INCLUDED, BUT THE TEMPERATURE FACTORS ARE VERY HIGH.
| ||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.8→9 Å
| ||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||
| Software | *PLUS Name: TNT / Classification: refinement | ||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller














 PDBj
PDBj







