[English] 日本語
 Yorodumi
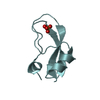
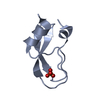
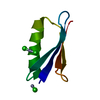
Yorodumi- PDB-1shp: THE NMR SOLUTION STRUCTURE OF A KUNITZ-TYPE PROTEINASE INHIBITOR ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1shp | ||||||
|---|---|---|---|---|---|---|---|
| Title | THE NMR SOLUTION STRUCTURE OF A KUNITZ-TYPE PROTEINASE INHIBITOR FROM THE SEA ANEMONE STICHODACTYLA HELIANTHUS | ||||||
 Components Components | TRYPSIN INHIBITOR | ||||||
 Keywords Keywords | PROTEINASE INHIBITOR(TRYPSIN) | ||||||
| Function / homology |  Function and homology information Function and homology information nematocyst / aspartic-type endopeptidase inhibitor activity / serine-type endopeptidase inhibitor activity / extracellular region nematocyst / aspartic-type endopeptidase inhibitor activity / serine-type endopeptidase inhibitor activity / extracellular regionSimilarity search - Function | ||||||
| Biological species |   Stichodactyla helianthus (sea anemone) Stichodactyla helianthus (sea anemone) | ||||||
| Method |  SOLUTION NMR SOLUTION NMR | ||||||
 Authors Authors | Antuch, W. / Berndt, K. / Chavez, M. / Delfin, J. / Wuthrich, K. | ||||||
 Citation Citation |  Journal: Eur.J.Biochem. / Year: 1993 Journal: Eur.J.Biochem. / Year: 1993Title: The NMR solution structure of a Kunitz-type proteinase inhibitor from the sea anemone Stichodactyla helianthus. Authors: Antuch, W. / Berndt, K.D. / Chavez, M.A. / Delfin, J. / Wuthrich, K. #1:  Journal: To be Published Journal: To be PublishedTitle: Amino Acid Sequence and Three-Dimensional Model of a Serine Proteinase Inhibitor from Stichodactyla Helianthus Authors: Antuch, W. / Dominguez, R. / Rodriguez, R. / Delfin, J. / Morera, V. / Diaz, J. / Chavez, M. / Dideberg, O. / Padron, G. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1shp.cif.gz 1shp.cif.gz | 355.5 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1shp.ent.gz pdb1shp.ent.gz | 313.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1shp.json.gz 1shp.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/sh/1shp https://data.pdbj.org/pub/pdb/validation_reports/sh/1shp ftp://data.pdbj.org/pub/pdb/validation_reports/sh/1shp ftp://data.pdbj.org/pub/pdb/validation_reports/sh/1shp | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: LYS 27 - CYS 28 MODEL 1 OMEGA =148.03 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 2: PRO 6 - LYS 7 MODEL 3 OMEGA =149.60 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 3: ARG 54 - ALA 55 MODEL 3 OMEGA =148.01 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 4: ASN 41 - ASN 42 MODEL 4 OMEGA =211.40 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 5: ARG 54 - ALA 55 MODEL 5 OMEGA =139.40 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 6: ARG 54 - ALA 55 MODEL 13 OMEGA =148.76 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 7: ASN 41 - ASN 42 MODEL 16 OMEGA =213.09 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 8: SER 1 - ILE 2 MODEL 18 OMEGA =212.95 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION 9: ARG 54 - ALA 55 MODEL 20 OMEGA =117.86 PEPTIDE BOND DEVIATES SIGNIFICANTLY FROM TRANS CONFORMATION | |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein |  Mass: 6124.990 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Stichodactyla helianthus (sea anemone) / References: UniProt: P31713 Stichodactyla helianthus (sea anemone) / References: UniProt: P31713 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR |
|---|
- Processing
Processing
| NMR software |
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| NMR ensemble | Conformers submitted total number: 20 |
 Movie
Movie Controller
Controller









 PDBj
PDBj



