+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1rl4 | ||||||
|---|---|---|---|---|---|---|---|
| Title | Plasmodium falciparum peptide deformylase complex with inhibitor | ||||||
 Components Components | formylmethionine deformylase | ||||||
 Keywords Keywords |  HYDROLASE / HYDROLASE /  crystal engineering / crystal engineering /  drug design / drug design /  malaria / PDF / malaria / PDF /  peptide deformylase / peptide deformylase /  Plasmodium Plasmodium | ||||||
| Function / homology |  Function and homology information Function and homology information apicoplast / co-translational protein modification / apicoplast / co-translational protein modification /  N-terminal protein amino acid modification / N-terminal protein amino acid modification /  peptide deformylase / peptide deformylase /  peptide deformylase activity / peptide deformylase activity /  ferrous iron binding / ferrous iron binding /  translation / translation /  mitochondrion / mitochondrion /  membrane membraneSimilarity search - Function | ||||||
| Biological species |   Plasmodium falciparum (malaria parasite P. falciparum) Plasmodium falciparum (malaria parasite P. falciparum) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / SYNCHROTRON /  MOLECULAR REPLACEMENT / Resolution: 2.18 Å MOLECULAR REPLACEMENT / Resolution: 2.18 Å | ||||||
 Authors Authors | Robien, M.A. / Nguyen, K.T. / Kumar, A. / Hirsh, I. / Turley, S. / Pei, D. / Hol, W.G.J. | ||||||
 Citation Citation |  Journal: Protein Sci. / Year: 2004 Journal: Protein Sci. / Year: 2004Title: An improved crystal form of Plasmodium falciparum peptide deformylase. Authors: Robien, M.A. / Nguyen, K.T. / Kumar, A. / Hirsh, I. / Turley, S. / Pei, D. / Hol, W.G.J. #1:  Journal: Structure / Year: 2002 Journal: Structure / Year: 2002Title: Crystals of Peptide Deformylase from Plasmodium falciparum Reveal Critical Characteristics of the Active Site for Drug Design Authors: Kumar, A. / Nguyen, K.T. / Srivathsan, S. / Ornstein, B. / Turley, S. / Hirsh, I. / Pei, D. / Hol, W.G.J. | ||||||
| History |
| ||||||
| Remark 600 | HETEROGEN AS DISCUSSED IN REFERENCE 1 (ROBIEN ET AL, PROTEIN SCIENCE), THE MODEL CHOSEN FOR BL5, ...HETEROGEN AS DISCUSSED IN REFERENCE 1 (ROBIEN ET AL, PROTEIN SCIENCE), THE MODEL CHOSEN FOR BL5, PARTICULARLY IN THE ATOMS LINKING THE TWO HALVES OF THIS MOLECULE, WAS CHOSEN BASED ON THE ELECTRON DENSITY FEATURE USING OMIT MAPS. FROM THE PROTEIN SCIENCE MANUSCRIPT: ALTHOUGH THE IDENTITY OF THE LINKER ATOMS ARE UNKNOWN, A LINEAR -CH2-NH-NH-CH2- CHAIN IS APPROXIMATELY THE CORRECT SIZE. BECAUSE OF THE UNCERTAINTY PERTAINING TO THE LINKER, THE MODEL CHOSEN FOR THE LINKER SHOULD BE CONSIDERED AS EXPLORATORY AND PROVISIONAL, RATHER THAN CONCLUSIVE. WATERS 701-753 AND 801-818, CO 301, BRR 401, AND BL5 501 ARE ASSOCIATED WITH CHAIN A. WATERS 1701-1753 AND 901-953, CO 1301, BRR 1401, AND BL5 1501 ARE ASSOCIATED WITH CHAIN B. | ||||||
| Remark 999 | SEQUENCE THE NUMBERING OF RESIDUES IS BASED ON THE FULL SEQUENCE OF THE P.FALCIPARUM PDF PROTEIN, ...SEQUENCE THE NUMBERING OF RESIDUES IS BASED ON THE FULL SEQUENCE OF THE P.FALCIPARUM PDF PROTEIN, WHICH HAS A LARGE N-TERMINAL (APICOPLAST TARGETING) SIGNAL SEQUENCE, WHICH IS TRUNCATED IN VIVO IN THE PROCESS OF TRANSPORT INTO THE APICOPLAST ORGANELLE. A CONSTRUCT WITH AN N-TERMINAL 57 RESIDUE TRUNCATION WAS EXPRESSED IN E COLI FOR THIS STUDY. THE C-TERMINAL SEQUENCE USED IN THIS STUDY DIFFERS FROM THAT USED FOR PDB ENTRY 1JYM BY THE DELETION OF THREE RESIDUES (-EEP-) JUST PRIOR TO THE -LEHHHHHH OF THE HEXAHISTADINE TAG. THIS MODIFICATION WAS UNDERTAKEN AS A SUCCESSFUL EFFORT OF CRYSTAL ENGINEERING TO AVOID A NETWORK OF UNDESIRABLE CRYSTAL CONTACTS IN THE PREVIOUS STUDY. THE SEQUENCE OF P. FALCIPARUM PDF IN THIS DEPOSITION INCLUDES TWO SINGLE AMINO ACID CORRECTIONS TO THE SEQUENCE REPORTED IN THE ORIGINAL DEPOSITION (1JYM) OF P. FALCIPARUM PDF. RESIDUES GLU81 AND PRO160 HAD BEEN INADVERTENTLY NOT INCLUDED IN THE SEQUENCE USED DURING THE TRACING OF THE ELECTRON DENSITY OF THE ORIGINAL STUDY. THE RESIDUE NUMBERING OF THIS DEPOSITION REFLECTS THE MODIFIED, CORRECTED SEQUENCE INFORMATION, AND NOW AGREES WITH THE SEQUENCE DATA DERIVED FROM THE DEPOSITED P. FALCIPARUM GENOME. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1rl4.cif.gz 1rl4.cif.gz | 89.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1rl4.ent.gz pdb1rl4.ent.gz | 65.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1rl4.json.gz 1rl4.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/rl/1rl4 https://data.pdbj.org/pub/pdb/validation_reports/rl/1rl4 ftp://data.pdbj.org/pub/pdb/validation_reports/rl/1rl4 ftp://data.pdbj.org/pub/pdb/validation_reports/rl/1rl4 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1rqcC  1jymS S: Starting model for refinement C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 2 | 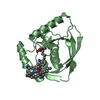
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Unit cell |
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Noncrystallographic symmetry (NCS) | NCS domain:
NCS domain segments:
|
 Movie
Movie Controller
Controller













 PDBj
PDBj