[English] 日本語
 Yorodumi
Yorodumi- PDB-1ft2: CO-CRYSTAL STRUCTURE OF PROTEIN FARNESYLTRANSFERASE COMPLEXED WIT... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ft2 | ||||||
|---|---|---|---|---|---|---|---|
| Title | CO-CRYSTAL STRUCTURE OF PROTEIN FARNESYLTRANSFERASE COMPLEXED WITH A FARNESYL DIPHOSPHATE SUBSTRATE | ||||||
 Components Components | (PROTEIN FARNESYLTRANSFERASE Farnesyltransferase) x 2 Farnesyltransferase) x 2 | ||||||
 Keywords Keywords |  TRANSFERASE / TRANSFERASE /  PROTEIN FARNESYLTRANSFERASE / PROTEIN FARNESYLTRANSFERASE /  FARNESYL DIPHOSPHATE / CANCER THERAPEUTICS / FARNESYL DIPHOSPHATE / CANCER THERAPEUTICS /  PRENYLTRANSFERASE / PRENYLTRANSFERASE /  ISOPRENOID ISOPRENOID | ||||||
| Function / homology |  Function and homology information Function and homology informationApoptotic cleavage of cellular proteins / Inactivation, recovery and regulation of the phototransduction cascade / RAS processing / protein geranylgeranyltransferase activity / CAAX-protein geranylgeranyltransferase activity / CAAX-protein geranylgeranyltransferase complex / protein farnesylation /  protein geranylgeranyltransferase type I / regulation of fibroblast proliferation / positive regulation of tubulin deacetylation ...Apoptotic cleavage of cellular proteins / Inactivation, recovery and regulation of the phototransduction cascade / RAS processing / protein geranylgeranyltransferase activity / CAAX-protein geranylgeranyltransferase activity / CAAX-protein geranylgeranyltransferase complex / protein farnesylation / protein geranylgeranyltransferase type I / regulation of fibroblast proliferation / positive regulation of tubulin deacetylation ...Apoptotic cleavage of cellular proteins / Inactivation, recovery and regulation of the phototransduction cascade / RAS processing / protein geranylgeranyltransferase activity / CAAX-protein geranylgeranyltransferase activity / CAAX-protein geranylgeranyltransferase complex / protein farnesylation /  protein geranylgeranyltransferase type I / regulation of fibroblast proliferation / positive regulation of tubulin deacetylation / protein geranylgeranyltransferase type I / regulation of fibroblast proliferation / positive regulation of tubulin deacetylation /  protein farnesyltransferase / protein farnesyltransferase /  protein farnesyltransferase activity / protein farnesyltransferase activity /  protein farnesyltransferase complex / protein farnesyltransferase complex /  Rab geranylgeranyltransferase activity / protein geranylgeranylation / positive regulation of skeletal muscle acetylcholine-gated channel clustering / Rab geranylgeranyltransferase activity / protein geranylgeranylation / positive regulation of skeletal muscle acetylcholine-gated channel clustering /  farnesyltranstransferase activity / negative regulation of nitric-oxide synthase biosynthetic process / microtubule associated complex / farnesyltranstransferase activity / negative regulation of nitric-oxide synthase biosynthetic process / microtubule associated complex /  enzyme-linked receptor protein signaling pathway / positive regulation of Rac protein signal transduction / positive regulation of nitric-oxide synthase biosynthetic process / alpha-tubulin binding / response to inorganic substance / positive regulation of cell cycle / response to cytokine / enzyme-linked receptor protein signaling pathway / positive regulation of Rac protein signal transduction / positive regulation of nitric-oxide synthase biosynthetic process / alpha-tubulin binding / response to inorganic substance / positive regulation of cell cycle / response to cytokine /  wound healing / lipid metabolic process / response to organic cyclic compound / wound healing / lipid metabolic process / response to organic cyclic compound /  receptor tyrosine kinase binding / positive regulation of fibroblast proliferation / fibroblast proliferation / receptor tyrosine kinase binding / positive regulation of fibroblast proliferation / fibroblast proliferation /  microtubule binding / cell population proliferation / molecular adaptor activity / negative regulation of cell population proliferation / positive regulation of cell population proliferation / negative regulation of apoptotic process / zinc ion binding / microtubule binding / cell population proliferation / molecular adaptor activity / negative regulation of cell population proliferation / positive regulation of cell population proliferation / negative regulation of apoptotic process / zinc ion binding /  cytoplasm cytoplasmSimilarity search - Function | ||||||
| Biological species |   Rattus norvegicus (Norway rat) Rattus norvegicus (Norway rat) | ||||||
| Method |  X-RAY DIFFRACTION / MOLECULAR REPLACEMENT USING 1FT1 COORDINATES / Resolution: 3.4 Å X-RAY DIFFRACTION / MOLECULAR REPLACEMENT USING 1FT1 COORDINATES / Resolution: 3.4 Å | ||||||
 Authors Authors | Beese, L.S. / Casey, P.J. / Long, S.B. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1998 Journal: Biochemistry / Year: 1998Title: Cocrystal structure of protein farnesyltransferase complexed with a farnesyl diphosphate substrate. Authors: Long, S.B. / Casey, P.J. / Beese, L.S. #1:  Journal: Science / Year: 1997 Journal: Science / Year: 1997Title: Crystal Structure of Protein Farnesyltransferase at 2.25 Angstrom Resolution Authors: Park, H.W. / Boduluri, S.R. / Moomaw, J.F. / Casey, P.J. / Beese, L.S. #2:  Journal: Science / Year: 1997 Journal: Science / Year: 1997Title: Erratum. Crystal Structure of Protein Farnesyltransferase at 2.25 Angstrom Resolution Authors: Park, H.W. / Boduluri, S.R. / Moomaw, J.F. / Casey, P.J. / Beese, L.S. #3:  Journal: J.Biol.Chem. / Year: 1993 Journal: J.Biol.Chem. / Year: 1993Title: High Level Expression of Mammalian Protein Farnesyltransferase in a Baculovirus System. The Purified Protein Contains Zinc Authors: Chen, W.J. / Moomaw, J.F. / Overton, L. / Kost, T.A. / Casey, P.J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ft2.cif.gz 1ft2.cif.gz | 151.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ft2.ent.gz pdb1ft2.ent.gz | 121.7 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ft2.json.gz 1ft2.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ft/1ft2 https://data.pdbj.org/pub/pdb/validation_reports/ft/1ft2 ftp://data.pdbj.org/pub/pdb/validation_reports/ft/1ft2 ftp://data.pdbj.org/pub/pdb/validation_reports/ft/1ft2 | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1ft1S S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  Farnesyltransferase FarnesyltransferaseMass: 38028.766 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Rattus norvegicus (Norway rat) / Cellular location: CYTOPLASM Rattus norvegicus (Norway rat) / Cellular location: CYTOPLASM / Gene: CDNA / Organ: BRAIN / Gene: CDNA / Organ: BRAIN / Plasmid: BACULOVIRUS / Cell line (production host): SF9 / Production host: / Plasmid: BACULOVIRUS / Cell line (production host): SF9 / Production host:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm)References: UniProt: Q04631,  Transferases; Transferring alkyl or aryl groups, other than methyl groups Transferases; Transferring alkyl or aryl groups, other than methyl groups |
|---|---|
| #2: Protein |  Farnesyltransferase FarnesyltransferaseMass: 44829.238 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Rattus norvegicus (Norway rat) / Cellular location: CYTOPLASM Rattus norvegicus (Norway rat) / Cellular location: CYTOPLASM / Gene: CDNA / Organ: BRAIN / Gene: CDNA / Organ: BRAIN / Plasmid: BACULOVIRUS / Cell line (production host): SF9 / Production host: / Plasmid: BACULOVIRUS / Cell line (production host): SF9 / Production host:   Spodoptera frugiperda (fall armyworm) Spodoptera frugiperda (fall armyworm)References: UniProt: Q02293,  Transferases; Transferring alkyl or aryl groups, other than methyl groups Transferases; Transferring alkyl or aryl groups, other than methyl groups |
| #3: Chemical | ChemComp-ZN / |
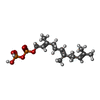
| #4: Chemical | ChemComp-FPP /  Farnesyl pyrophosphate Farnesyl pyrophosphate |
| Compound details | THIS PDB ENTRY CONTAINS THE COORDINATES OF A 3.4 ANGSTROM RESOLUTION CO-CRYSTAL STRUCTURE OF ...THIS PDB ENTRY CONTAINS THE COORDINATE |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.79 Å3/Da / Density % sol: 68 % | |||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | Method: vapor diffusion, hanging drop / pH: 7 Details: HANGING DROP, PROTEIN CONCENTRATION OF 16MG/ML IN 20 MM KCL, 10UM ZNCL, 10MM DTT, 20MM TRIS PH 7.7, 0.12% OCTYL-BETA-D-GLUCOPYRANOSIDE, AND 1MM FARNESYL DIPHOSPHATE; RESERVOIR SOLUTION OF ...Details: HANGING DROP, PROTEIN CONCENTRATION OF 16MG/ML IN 20 MM KCL, 10UM ZNCL, 10MM DTT, 20MM TRIS PH 7.7, 0.12% OCTYL-BETA-D-GLUCOPYRANOSIDE, AND 1MM FARNESYL DIPHOSPHATE; RESERVOIR SOLUTION OF 15% PEG 8000, 200MM AMMONIUM ACETATE, PH 7.0, vapor diffusion - hanging drop PH range: 7.0-7.7 | |||||||||||||||||||||||||||||||||||||||||||||
| Crystal | *PLUS | |||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 17 ℃ / pH: 7.7 / Method: vapor diffusion, hanging drop | |||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 96 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU / Wavelength: 1.5418 |
| Detector | Type: RIGAKU / Detector: IMAGE PLATE / Date: Apr 27, 1998 / Details: MSC DOUBLE-MIRRORS |
| Radiation | Monochromator: NI FILTER / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 3.4→35 Å / Num. obs: 17125 / % possible obs: 79.4 % / Observed criterion σ(I): 0 / Redundancy: 2.5 % / Rmerge(I) obs: 0.053 / Rsym value: 0.053 / Net I/σ(I): 15 |
| Reflection shell | Resolution: 3.4→3.44 Å / Redundancy: 1.8 % / Rmerge(I) obs: 0.199 / Mean I/σ(I) obs: 2.7 / Rsym value: 0.199 / % possible all: 56.1 |
| Reflection | *PLUS Num. measured all: 42609 |
| Reflection shell | *PLUS % possible obs: 58.1 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : MOLECULAR REPLACEMENT USING 1FT1 COORDINATES : MOLECULAR REPLACEMENT USING 1FT1 COORDINATESStarting model: PDB ENTRY 1FT1 Resolution: 3.4→35 Å / Rfactor Rfree error: 0.009 / Data cutoff high absF: 10000000 / Data cutoff low absF: 0.001 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 Details: THE CRYSTAL STRUCTURE OF PROTEIN FARNESYLTRANSFERASE WITHOUT SUBSTRATE BOUND WAS DETERMINED TO 2.25 ANGSTROM RESOLUTION (APO FTASE, PDB ID CODE: 1FT1). PHASES FOR THE CO-CRYSTAL STRUCTURE OF ...Details: THE CRYSTAL STRUCTURE OF PROTEIN FARNESYLTRANSFERASE WITHOUT SUBSTRATE BOUND WAS DETERMINED TO 2.25 ANGSTROM RESOLUTION (APO FTASE, PDB ID CODE: 1FT1). PHASES FOR THE CO-CRYSTAL STRUCTURE OF PROTEIN FARNESYLTRANSFERASE WITH FARNESYL DIPHOSPHATE BOUND WERE DERIVED FROM THIS HIGH RESOLUTION APO FTASE CRYSTAL STRUCTURE.
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | Biso mean: 40.4 Å2 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 3.4→35 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 3.4→3.55 Å / Rfactor Rfree error: 0.036 / Total num. of bins used: 8
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name:  X-PLOR / Version: 3.851 / Classification: refinement X-PLOR / Version: 3.851 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor Rfree: 0.32 |
 Movie
Movie Controller
Controller




















 PDBj
PDBj