[English] 日本語
 Yorodumi
Yorodumi- PDB-1ezv: STRUCTURE OF THE YEAST CYTOCHROME BC1 COMPLEX CO-CRYSTALLIZED WIT... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1ezv | ||||||
|---|---|---|---|---|---|---|---|
| Title | STRUCTURE OF THE YEAST CYTOCHROME BC1 COMPLEX CO-CRYSTALLIZED WITH AN ANTIBODY FV-FRAGMENT | ||||||
 Components Components |
| ||||||
 Keywords Keywords | OXIDOREDUCTASE/ELECTRON TRANSPORT /  cytochrome bc1 complex / cytochrome bc1 complex /  complex III / QCR / complex III / QCR /  mitochondria / mitochondria /  yeast / antibody Fv-fragment / yeast / antibody Fv-fragment /  stigmatellin / coenzyme Q6 / matrix processing peptidases / stigmatellin / coenzyme Q6 / matrix processing peptidases /  ubiquinone / ubiquinone /  electron transfer / proton transfer / Q-cycle / OXIDOREDUCTASE-ELECTRON TRANSPORT COMPLEX electron transfer / proton transfer / Q-cycle / OXIDOREDUCTASE-ELECTRON TRANSPORT COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationScavenging of heme from plasma / Fc epsilon receptor (FCERI) signaling / CD22 mediated BCR regulation / Role of LAT2/NTAL/LAB on calcium mobilization / FCERI mediated MAPK activation / Initial triggering of complement / Classical antibody-mediated complement activation / FCGR activation / Role of phospholipids in phagocytosis /  Regulation of Complement cascade ...Scavenging of heme from plasma / Fc epsilon receptor (FCERI) signaling / CD22 mediated BCR regulation / Role of LAT2/NTAL/LAB on calcium mobilization / FCERI mediated MAPK activation / Initial triggering of complement / Classical antibody-mediated complement activation / FCGR activation / Role of phospholipids in phagocytosis / Regulation of Complement cascade ...Scavenging of heme from plasma / Fc epsilon receptor (FCERI) signaling / CD22 mediated BCR regulation / Role of LAT2/NTAL/LAB on calcium mobilization / FCERI mediated MAPK activation / Initial triggering of complement / Classical antibody-mediated complement activation / FCGR activation / Role of phospholipids in phagocytosis /  Regulation of Complement cascade / matrix side of mitochondrial inner membrane / FCERI mediated Ca+2 mobilization / protein processing involved in protein targeting to mitochondrion / FCERI mediated NF-kB activation / Cell surface interactions at the vascular wall / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / Respiratory electron transport / Regulation of actin dynamics for phagocytic cup formation / mitochondrial respiratory chain complex III assembly / mitochondrial respiratory chain complex III / quinol-cytochrome-c reductase / Regulation of Complement cascade / matrix side of mitochondrial inner membrane / FCERI mediated Ca+2 mobilization / protein processing involved in protein targeting to mitochondrion / FCERI mediated NF-kB activation / Cell surface interactions at the vascular wall / Antigen activates B Cell Receptor (BCR) leading to generation of second messengers / Respiratory electron transport / Regulation of actin dynamics for phagocytic cup formation / mitochondrial respiratory chain complex III assembly / mitochondrial respiratory chain complex III / quinol-cytochrome-c reductase /  cellular respiration / cellular respiration /  ubiquinol-cytochrome-c reductase activity / mitochondrial electron transport, ubiquinol to cytochrome c / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell / ubiquinol-cytochrome-c reductase activity / mitochondrial electron transport, ubiquinol to cytochrome c / Immunoregulatory interactions between a Lymphoid and a non-Lymphoid cell /  immunoglobulin complex / mitochondrial crista / immunoglobulin mediated immune response / immunoglobulin complex / mitochondrial crista / immunoglobulin mediated immune response /  aerobic respiration / aerobic respiration /  antigen binding / proton transmembrane transport / nuclear periphery / antigen binding / proton transmembrane transport / nuclear periphery /  mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding / mitochondrial intermembrane space / 2 iron, 2 sulfur cluster binding /  metalloendopeptidase activity / metalloendopeptidase activity /  mitochondrial inner membrane / mitochondrial inner membrane /  adaptive immune response / adaptive immune response /  oxidoreductase activity / oxidoreductase activity /  immune response / immune response /  heme binding / heme binding /  mitochondrion / mitochondrion /  proteolysis / proteolysis /  extracellular space / extracellular region / extracellular space / extracellular region /  metal ion binding / metal ion binding /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)  Mus musculus (house mouse) Mus musculus (house mouse) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 2.3 Å SYNCHROTRON / Resolution: 2.3 Å | ||||||
 Authors Authors | Hunte, C. / Koepke, J. / Lange, C. / Rossmanith, T. / Michel, H. | ||||||
 Citation Citation |  Journal: Structure Fold.Des. / Year: 2000 Journal: Structure Fold.Des. / Year: 2000Title: Structure at 2.3 A resolution of the cytochrome bc(1) complex from the yeast Saccharomyces cerevisiae co-crystallized with an antibody Fv fragment. Authors: Hunte, C. / Koepke, J. / Lange, C. / Rossmanith, T. / Michel, H. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1ezv.cif.gz 1ezv.cif.gz | 451.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1ezv.ent.gz pdb1ezv.ent.gz | 370.5 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1ezv.json.gz 1ezv.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ez/1ezv https://data.pdbj.org/pub/pdb/validation_reports/ez/1ezv ftp://data.pdbj.org/pub/pdb/validation_reports/ez/1ezv ftp://data.pdbj.org/pub/pdb/validation_reports/ez/1ezv | HTTPS FTP |
|---|
-Related structure data
| Related structure data | |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | 
| ||||||||
| Unit cell |
| ||||||||
| Details | The yeast mitochondrial cytochrome bc1 complex consist of 9 subunits (COR1, QCR2, COB, CYT1, RIP1, QCR6, QCR7, QCR8, QCR9). The biological functional unit is a homodimer. The smallest subunit QCR10, which is not required for a functional enzyme, was not present in the protein preparations. / The cytochrome bc1 complex was co-crystallized with an antibody Fv-fragment, which is bound to subunit RIP1 ("Rieske"-protein). The Fv-fragment consists of heavy and light chain (VH and VL). the Fv-fragment is bound to the "Rieske"-protein (chain ID E) |
- Components
Components
-UBIQUINOL-CYTOCHROME C REDUCTASE COMPLEX ... , 6 types, 6 molecules ABHFGI
| #1: Protein | Mass: 47358.168 Da / Num. of mol.: 1 / Fragment: RESIDUES 24-457 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Description: MITOCHONDRIA, YEAST, SACCHAROMYCES CEREVISIAE / Organelle: MITOCHONDRIA  Mitochondrion / References: UniProt: P07256, quinol-cytochrome-c reductase Mitochondrion / References: UniProt: P07256, quinol-cytochrome-c reductase |
|---|---|
| #2: Protein | Mass: 38751.918 Da / Num. of mol.: 1 / Fragment: RESIDUES 17-368 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Description: FV-FRAGMENT DERIVED FROM THE MURINE MONOCLONAL ANTIBODY 18E11, EXPRESSION SYSTEM ESCHERICHIA COLI Organelle: MITOCHONDRIA  Mitochondrion MitochondrionReferences:  GenBank: 786302, UniProt: P07257*PLUS, quinol-cytochrome-c reductase GenBank: 786302, UniProt: P07257*PLUS, quinol-cytochrome-c reductase |
| #6: Protein | Mass: 8854.792 Da / Num. of mol.: 1 / Fragment: RESIDUES 74-147 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Organelle: MITOCHONDRIA  Mitochondrion MitochondrionReferences:  GenBank: 836788, UniProt: P00127*PLUS, quinol-cytochrome-c reductase GenBank: 836788, UniProt: P00127*PLUS, quinol-cytochrome-c reductase |
| #7: Protein | Mass: 14355.443 Da / Num. of mol.: 1 / Fragment: RESIDUES 3-127 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Organelle: MITOCHONDRIA  Mitochondrion MitochondrionReferences:  GenBank: 927796, UniProt: P00128*PLUS, quinol-cytochrome-c reductase GenBank: 927796, UniProt: P00128*PLUS, quinol-cytochrome-c reductase |
| #8: Protein | Mass: 10856.314 Da / Num. of mol.: 1 / Fragment: RESIDUES 2-94 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Organelle: MITOCHONDRIA  Mitochondrion MitochondrionReferences:  GenBank: 1008356, UniProt: P08525*PLUS, quinol-cytochrome-c reductase GenBank: 1008356, UniProt: P08525*PLUS, quinol-cytochrome-c reductase |
| #9: Protein | Mass: 6301.232 Da / Num. of mol.: 1 / Fragment: RESIDUES 4-58 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Organelle: MITOCHONDRIA  Mitochondrion / References: UniProt: P22289, quinol-cytochrome-c reductase Mitochondrion / References: UniProt: P22289, quinol-cytochrome-c reductase |
-Protein , 3 types, 3 molecules CDE
| #3: Protein |  Mass: 43674.535 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Organelle: MITOCHONDRIA  Mitochondrion / References: Mitochondrion / References:  GenBank: 643021, UniProt: P00163*PLUS GenBank: 643021, UniProt: P00163*PLUS |
|---|---|
| #4: Protein |  Mass: 27423.904 Da / Num. of mol.: 1 / Fragment: RESIDUES 62-306 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Organelle: MITOCHONDRIA  Mitochondrion / References: Mitochondrion / References:  GenBank: 1420211, UniProt: P07143*PLUS GenBank: 1420211, UniProt: P07143*PLUS |
| #5: Protein | Mass: 20122.955 Da / Num. of mol.: 1 / Fragment: RESIDUES 31-215 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Saccharomyces cerevisiae (brewer's yeast) Saccharomyces cerevisiae (brewer's yeast)Organelle: MITOCHONDRIA  Mitochondrion MitochondrionReferences:  GenBank: 602391, UniProt: P08067*PLUS, quinol-cytochrome-c reductase GenBank: 602391, UniProt: P08067*PLUS, quinol-cytochrome-c reductase |
-Antibody , 2 types, 2 molecules XY
| #10: Antibody | Mass: 14365.817 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Production host: Mus musculus (house mouse) / Production host:   Escherichia coli (E. coli) / References: UniProt: P18531*PLUS Escherichia coli (E. coli) / References: UniProt: P18531*PLUS |
|---|---|
| #11: Antibody | Mass: 11926.350 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Production host: Mus musculus (house mouse) / Production host:   Escherichia coli (E. coli) / References: UniProt: P01647*PLUS Escherichia coli (E. coli) / References: UniProt: P01647*PLUS |
-Non-polymers , 5 types, 352 molecules 
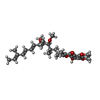







| #12: Chemical |  Heme B Heme B#13: Chemical | ChemComp-SMA / | #14: Chemical | ChemComp-UQ6 / | #15: Chemical | ChemComp-FES / |  Iron–sulfur cluster Iron–sulfur cluster#16: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 17 X-RAY DIFFRACTION / Number of used crystals: 17 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 4.7 Å3/Da / Density % sol: 73.85 % | ||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | Temperature: 277 K / Method: microseeding / pH: 8 Details: 5 % PEG 4000, 100 mM Tris, 0.05 % Undecyl-maltoside, 1 micromolar stigmatellin, pH 8.0, microseeding, temperature 277K | ||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Method: vapor diffusion | ||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diffraction source |
| |||||||||||||||
| Detector |
| |||||||||||||||
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray | |||||||||||||||
| Radiation wavelength |
| |||||||||||||||
| Reflection | Resolution: 2.3→15 Å / Num. all: 168625 / % possible obs: 84.9 % / Observed criterion σ(F): 3 / Observed criterion σ(I): 0 / Redundancy: 6.27 % / Biso Wilson estimate: 52.1 Å2 / Rmerge(I) obs: 0.065 / Net I/σ(I): 12.4 | |||||||||||||||
| Reflection shell | Resolution: 2.3→15 Å / Redundancy: 2.2 % / Rmerge(I) obs: 0.39 / Num. unique all: 4845 / % possible all: 73.9 | |||||||||||||||
| Reflection | *PLUS Lowest resolution: 15 Å / Num. obs: 168625 / Num. measured all: 1057968 | |||||||||||||||
| Reflection shell | *PLUS % possible obs: 73.9 % |
- Processing
Processing
| Software |
| |||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2.3→15 Å / σ(F): 0 / σ(I): 0 / Stereochemistry target values: Engh & Huber
| |||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.3→15 Å
| |||||||||||||||||||||||||
| Refine LS restraints |
| |||||||||||||||||||||||||
| Software | *PLUS Name: CNS / Classification: refinement | |||||||||||||||||||||||||
| Refinement | *PLUS Highest resolution: 2.3 Å / Lowest resolution: 15 Å / σ(F): 0 / % reflection Rfree: 2.5 % / Rfactor all : 0.222 : 0.222 | |||||||||||||||||||||||||
| Solvent computation | *PLUS | |||||||||||||||||||||||||
| Displacement parameters | *PLUS | |||||||||||||||||||||||||
| Refine LS restraints | *PLUS Type: c_angle_deg / Dev ideal: 0.9 |
 Movie
Movie Controller
Controller
















 PDBj
PDBj























