[English] 日本語
 Yorodumi
Yorodumi- PDB-1d3g: HUMAN DIHYDROOROTATE DEHYDROGENASE COMPLEXED WITH BREQUINAR ANALOG -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1d3g | ||||||
|---|---|---|---|---|---|---|---|
| Title | HUMAN DIHYDROOROTATE DEHYDROGENASE COMPLEXED WITH BREQUINAR ANALOG | ||||||
 Components Components | DIHYDROOROTATE DEHYDROGENASE | ||||||
 Keywords Keywords |  OXIDOREDUCTASE / PROTEIN-ANTIPROLIFERATIVE AGENT COMPLEX OXIDOREDUCTASE / PROTEIN-ANTIPROLIFERATIVE AGENT COMPLEX | ||||||
| Function / homology |  Function and homology information Function and homology informationpyrimidine ribonucleotide biosynthetic process /  dihydroorotate dehydrogenase (quinone) activity / dihydroorotate dehydrogenase (quinone) activity /  dihydroorotate dehydrogenase (quinone) / dihydroorotate dehydrogenase (quinone) /  dihydroorotate dehydrogenase activity / dihydroorotate dehydrogenase activity /  dihydroorotase activity / dihydroorotase activity /  Pyrimidine biosynthesis / UDP biosynthetic process / 'de novo' UMP biosynthetic process / 'de novo' pyrimidine nucleobase biosynthetic process / Pyrimidine biosynthesis / UDP biosynthetic process / 'de novo' UMP biosynthetic process / 'de novo' pyrimidine nucleobase biosynthetic process /  mitochondrial inner membrane ...pyrimidine ribonucleotide biosynthetic process / mitochondrial inner membrane ...pyrimidine ribonucleotide biosynthetic process /  dihydroorotate dehydrogenase (quinone) activity / dihydroorotate dehydrogenase (quinone) activity /  dihydroorotate dehydrogenase (quinone) / dihydroorotate dehydrogenase (quinone) /  dihydroorotate dehydrogenase activity / dihydroorotate dehydrogenase activity /  dihydroorotase activity / dihydroorotase activity /  Pyrimidine biosynthesis / UDP biosynthetic process / 'de novo' UMP biosynthetic process / 'de novo' pyrimidine nucleobase biosynthetic process / Pyrimidine biosynthesis / UDP biosynthetic process / 'de novo' UMP biosynthetic process / 'de novo' pyrimidine nucleobase biosynthetic process /  mitochondrial inner membrane / mitochondrial inner membrane /  mitochondrion / mitochondrion /  nucleoplasm / nucleoplasm /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  SYNCHROTRON / Resolution: 1.6 Å SYNCHROTRON / Resolution: 1.6 Å | ||||||
 Authors Authors | Liu, S. / Neidhardt, E.A. / Grossman, T.H. / Ocain, T. / Clardy, J. | ||||||
 Citation Citation |  Journal: Structure Fold.Des. / Year: 2000 Journal: Structure Fold.Des. / Year: 2000Title: Structures of human dihydroorotate dehydrogenase in complex with antiproliferative agents. Authors: Liu, S. / Neidhardt, E.A. / Grossman, T.H. / Ocain, T. / Clardy, J. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1d3g.cif.gz 1d3g.cif.gz | 165.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1d3g.ent.gz pdb1d3g.ent.gz | 129.4 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1d3g.json.gz 1d3g.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/d3/1d3g https://data.pdbj.org/pub/pdb/validation_reports/d3/1d3g ftp://data.pdbj.org/pub/pdb/validation_reports/d3/1d3g ftp://data.pdbj.org/pub/pdb/validation_reports/d3/1d3g | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
-Protein , 1 types, 1 molecules A
| #1: Protein |  Mass: 39856.535 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Plasmid: PET19B / Production host: Homo sapiens (human) / Plasmid: PET19B / Production host:   Escherichia coli (E. coli) / References: UniProt: Q02127, EC: 1.3.3.1 Escherichia coli (E. coli) / References: UniProt: Q02127, EC: 1.3.3.1 |
|---|
-Non-polymers , 7 types, 280 molecules 









| #2: Chemical | ChemComp-SO4 /  Sulfate Sulfate |
|---|---|
| #3: Chemical | ChemComp-ACT /  Acetate Acetate |
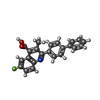
| #4: Chemical | ChemComp-BRE / |
| #5: Chemical | ChemComp-FMN /  Flavin mononucleotide Flavin mononucleotide |
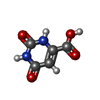
| #6: Chemical | ChemComp-ORO /  Orotic acid Orotic acid |
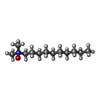
| #7: Chemical | ChemComp-DDQ / |
| #8: Water | ChemComp-HOH /  Water Water |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 1 X-RAY DIFFRACTION / Number of used crystals: 1 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 3.64 Å3/Da / Density % sol: 66.19 % | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | Temperature: 293 K / Method: vapor diffusion Details: AMMONIUM SULFATE, ACETATE, GLYCEROL, DDAO, C11DAO, BREQUINAR, OROTATE, VAPOR DIFFUSION, temperature 293K | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Crystal grow | *PLUS Temperature: 20 ℃ / Method: vapor diffusion, hanging drop | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 100 K |
|---|---|
| Diffraction source | Source:  SYNCHROTRON / Site: SYNCHROTRON / Site:  CHESS CHESS  / Beamline: A1 / Wavelength: 0.936 / Beamline: A1 / Wavelength: 0.936 |
| Detector | Type: ADSC QUANTUM 4 / Detector: CCD / Date: Dec 5, 1998 |
| Radiation | Protocol: SINGLE WAVELENGTH / Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 0.936 Å / Relative weight: 1 : 0.936 Å / Relative weight: 1 |
| Reflection | Resolution: 1.6→30 Å / Num. all: 77790 / Num. obs: 75457 / % possible obs: 97 % / Observed criterion σ(F): 0 / Observed criterion σ(I): 0 / Redundancy: 5.9 % / Biso Wilson estimate: 18.5 Å2 / Rmerge(I) obs: 0.04 / Net I/σ(I): 13 |
| Reflection shell | Resolution: 1.6→1.7 Å / Redundancy: 3.2 % / Rmerge(I) obs: 0.21 / % possible all: 83.5 |
| Reflection | *PLUS % possible obs: 97.1 % / Num. measured all: 444987 |
| Reflection shell | *PLUS % possible obs: 84.8 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 1.6→30 Å / Rfactor Rfree error: 0.003 / Isotropic thermal model: RESTRAINED / Cross valid method: THROUGHOUT / σ(F): 0 / σ(I): 0 / Stereochemistry target values: ENGH & HUBER / Details: USED WEIGHTED FULL MATRIX LEAST SQUARES PROCEDURE
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine analyze |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 1.6→30 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | Resolution: 1.6→1.7 Å / Rfactor Rfree error: 0.01 / Total num. of bins used: 6
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Xplor file |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: REFMAC / Version: 4 / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Lowest resolution: 30 Å / σ(F): 0 / % reflection Rfree: 5 % / Rfactor obs: 0.169 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| LS refinement shell | *PLUS Rfactor obs: 0.166 |
 Movie
Movie Controller
Controller
















 PDBj
PDBj






