[English] 日本語
 Yorodumi
Yorodumi- PDB-1cya: NMR STUDIES OF (U-13C)CYCLOSPORIN A BOUND TO CYCLOPHILIN: BOUND C... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1cya | ||||||
|---|---|---|---|---|---|---|---|
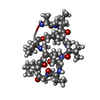
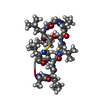
| Title | NMR STUDIES OF (U-13C)CYCLOSPORIN A BOUND TO CYCLOPHILIN: BOUND CONFORMATION AND PORTIONS OF CYCLOSPORIN INVOLVED IN BINDING | ||||||
 Components Components | CYCLOSPORIN A Ciclosporin Ciclosporin | ||||||
 Keywords Keywords |  IMMUNOSUPPRESSANT / IMMUNOSUPPRESSANT /  CYCLOSPORIN A CYCLOSPORIN A | ||||||
| Function / homology |  Cyclosporin A / Cyclosporin A /  : :  Function and homology information Function and homology information | ||||||
| Biological species |   TOLYPOCLADIUM INFLATUM (fungus) TOLYPOCLADIUM INFLATUM (fungus) | ||||||
| Method |  SOLUTION NMR SOLUTION NMR | ||||||
| Model type details | MINIMIZED AVERAGE | ||||||
 Authors Authors | Fesik, S.W. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1991 Journal: Biochemistry / Year: 1991Title: NMR Studies of [U-13C]Cyclosporin a Bound to Cyclophilin: Bound Conformation and Portions of Cyclosporin Involved in Binding. Authors: Fesik, S.W. / Gampe, R.T. / Eaton, H.L. / Gemmecker, G. / Olejniczak, E.T. / Neri, P. / Holzman, T.F. / Egan, D.A. / Edalji, R. #1:  Journal: J.Am.Chem.Soc. / Year: 1991 Journal: J.Am.Chem.Soc. / Year: 1991Title: Identification of Solvent-Exposed Regions of Enzyme-Bound Ligands by Nuclear Magnetic Resonance Authors: Fesik, S.W. / Gemmecker, G. / Olejniczak, E.T. / Petros, A.M. #2:  Journal: FEBS Lett. / Year: 1991 Journal: FEBS Lett. / Year: 1991Title: 1H, 13C and 15N Backbone Assignments of Cyclophilin When Bound to Cyclosporin a (Csa) and Preliminary Structural Characterization of the Csa Binding Site Authors: Neri, P. / Meadows, R. / Gemmecker, G. / Olejniczak, E. / Nettesheim, D. / Logan, T. / Simmer, R. / Helfrich, R. / Holzman, T. / Severin, J. / Fesik, S. | ||||||
| History |
|
- Structure visualization
Structure visualization
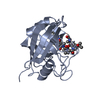
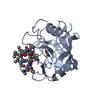
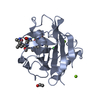
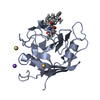
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1cya.cif.gz 1cya.cif.gz | 9.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1cya.ent.gz pdb1cya.ent.gz | 8.2 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1cya.json.gz 1cya.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/cy/1cya https://data.pdbj.org/pub/pdb/validation_reports/cy/1cya ftp://data.pdbj.org/pub/pdb/validation_reports/cy/1cya ftp://data.pdbj.org/pub/pdb/validation_reports/cy/1cya | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| Atom site foot note | 1: RESIDUE ALA 8 IS A D-ALANINE. | |||||||||
| NMR ensembles |
|
- Components
Components
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR |
|---|
- Sample preparation
Sample preparation
Crystal grow | *PLUS Method: other / Details: NMR |
|---|
- Processing
Processing
| Software |
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| NMR software | Name: DSPACE,  X-PLOR / Developer: HARE RESEARCH INC. (DSPACE), BRUNGER (X-PLOR) / Classification: refinement X-PLOR / Developer: HARE RESEARCH INC. (DSPACE), BRUNGER (X-PLOR) / Classification: refinement | ||||||||
| NMR ensemble | Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller




































 PDBj
PDBj