+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1c4l | ||||||
|---|---|---|---|---|---|---|---|
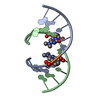
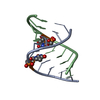
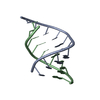
| Title | SOLUTION STRUCTURE OF AN RNA DUPLEX INCLUDING A C-U BASE-PAIR | ||||||
 Components Components |
| ||||||
 Keywords Keywords |  RNA / C-U base pair RNA / C-U base pair | ||||||
| Function / homology |  RNA RNA Function and homology information Function and homology information | ||||||
| Method |  SOLUTION NMR / SOLUTION NMR /  simulated annealing simulated annealing | ||||||
| Model type details | minimized average | ||||||
 Authors Authors | Tanaka, Y. / Kojima, C. / Yamazaki, T. / Kodama, T.S. / Yasuno, K. / Miyashita, S. / Ono, A.M. / Ono, A.S. / Kainosho, M. / Kyogoku, Y. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 2000 Journal: Biochemistry / Year: 2000Title: Solution structure of an RNA duplex including a C-U base pair. Authors: Tanaka, Y. / Kojima, C. / Yamazaki, T. / Kodama, T.S. / Yasuno, K. / Miyashita, S. / Ono, A. / Ono, A. / Kainosho, M. / Kyogoku, Y. #1:  Journal: Nucleic Acids Symp.Ser. / Year: 1997 Journal: Nucleic Acids Symp.Ser. / Year: 1997Title: Structure model and physicochemical properties of the C-U mismatch pair in the double stranded RNA in solution Authors: Tanaka, Y. / Kojima, C. / Yamazaki, T. / Kyogoku, Y. / Miyashita, S. / Ono, A.S. / Kainosho, M. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1c4l.cif.gz 1c4l.cif.gz | 19.6 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1c4l.ent.gz pdb1c4l.ent.gz | 12.3 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1c4l.json.gz 1c4l.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/c4/1c4l https://data.pdbj.org/pub/pdb/validation_reports/c4/1c4l ftp://data.pdbj.org/pub/pdb/validation_reports/c4/1c4l ftp://data.pdbj.org/pub/pdb/validation_reports/c4/1c4l | HTTPS FTP |
|---|
-Related structure data
| Similar structure data |
|---|
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: RNA chain | Mass: 2870.783 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: THIS OLIGONUCLEOTIDE WAS CHEMICALLY SYNTHESIZED (PHOSPHORAMIDITE METHOD). |
|---|---|
| #2: RNA chain | Mass: 2823.719 Da / Num. of mol.: 1 / Source method: obtained synthetically Details: THIS OLIGONUCLEOTIDE WAS CHEMICALLY SYNTHESIZED (PHOSPHORAMIDITE METHOD). |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||||||
| NMR details | Text: THIS STRUCTURE WAS DETERMINED USING STANDARD 2D HOMONUCLEAR TECHNIQUES |
- Sample preparation
Sample preparation
| Details |
| |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample conditions |
| |||||||||||||||
Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method:  simulated annealing / Software ordinal: 1 simulated annealing / Software ordinal: 1 Details: DISTANCE CONSTRAINTS (NOE 433, HYDROGEN BOND (BASE-PAIR) 54), DIHEDRAL ANGLE CONSTRAINTS 45 | ||||||||||||||||||||
| NMR representative | Selection criteria: minimized average structure | ||||||||||||||||||||
| NMR ensemble | Conformer selection criteria: SUMMATION OF TOTAL ENERGY (COVALENT GEOMETRY, NON-BOND ENERGY, DISTANCE AND DIHEDRAL ANGLE CONSTRAINTS) Conformers calculated total number: 50 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller




 PDBj
PDBj