[English] 日本語
 Yorodumi
Yorodumi- PDB-1b3i: NMR SOLUTION STRUCTURE OF PLASTOCYANIN FROM THE PHOTOSYNTHETIC PR... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1b3i | ||||||
|---|---|---|---|---|---|---|---|
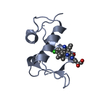
| Title | NMR SOLUTION STRUCTURE OF PLASTOCYANIN FROM THE PHOTOSYNTHETIC PROKARYOTE, PROCHLOROTHRIX HOLLANDICA (MINIMIZED AVERAGE STRUCTURE) | ||||||
 Components Components | PROTEIN (PLASTOCYANIN) | ||||||
 Keywords Keywords |  ELECTRON TRANSPORT / TYPE I COPPER PROTEIN / ELECTRON TRANSPORT / TYPE I COPPER PROTEIN /  PHOTOSYNTHESIS PHOTOSYNTHESIS | ||||||
| Function / homology |  Function and homology information Function and homology informationplasma membrane-derived thylakoid membrane /  electron transfer activity / copper ion binding electron transfer activity / copper ion bindingSimilarity search - Function | ||||||
| Biological species |  Prochlorothrix hollandica (bacteria) Prochlorothrix hollandica (bacteria) | ||||||
| Method |  SOLUTION NMR / torsion angle dynamics SOLUTION NMR / torsion angle dynamics | ||||||
 Authors Authors | Babu, C.R. / Volkman, B.F. / Bullerjahn, G.S. | ||||||
 Citation Citation |  Journal: Biochemistry / Year: 1999 Journal: Biochemistry / Year: 1999Title: NMR solution structure of plastocyanin from the photosynthetic prokaryote, Prochlorothrix hollandica. Authors: Babu, C.R. / Volkman, B.F. / Bullerjahn, G.S. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1b3i.cif.gz 1b3i.cif.gz | 40.2 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1b3i.ent.gz pdb1b3i.ent.gz | 27.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1b3i.json.gz 1b3i.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/b3/1b3i https://data.pdbj.org/pub/pdb/validation_reports/b3/1b3i ftp://data.pdbj.org/pub/pdb/validation_reports/b3/1b3i ftp://data.pdbj.org/pub/pdb/validation_reports/b3/1b3i | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein | Mass: 10148.560 Da / Num. of mol.: 1 / Mutation: T2S Source method: isolated from a genetically manipulated source Details: T2S MUTATION INTRODUCED TO CLONE IN EXPRESSION VECTOR Source: (gene. exp.)  Prochlorothrix hollandica (bacteria) / Description: INCLUSION BODIES WERE REFOLDED IN-VITRO / Cellular location: THYLAKOID LUMEN Prochlorothrix hollandica (bacteria) / Description: INCLUSION BODIES WERE REFOLDED IN-VITRO / Cellular location: THYLAKOID LUMEN Thylakoid / Gene: PETE / Plasmid: PVAPC10 Thylakoid / Gene: PETE / Plasmid: PVAPC10Cellular location (production host): CYTOPLASM - INCLUSION BODIES Gene (production host): PETE / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21(DE3)PLYSS / References: UniProt: P50057, EC: 1.10.99.1 Escherichia coli (E. coli) / Strain (production host): BL21(DE3)PLYSS / References: UniProt: P50057, EC: 1.10.99.1 |
|---|---|
| #2: Chemical | ChemComp-CU1 /  Copper Copper |
| Sequence details | T2S - INTRODUCED |
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
| ||||||||||||||||
| NMR details | Text: MINIMIZED AVERAGE STRUCTURE. THE STRUCTURE WAS DETERMINED USING TWO- DIMENCIONAL NMR SPECTROSCOPY. WATER SUPPRESSION WAS ACHIEVED WITH A WATERGATE SEQ. WITH A 3-9-19 SELECTIVE INVERSION. |
- Sample preparation
Sample preparation
| Details | Contents: 90% WATER/10% D2O |
|---|---|
| Sample conditions | Ionic strength: 20mM POTASSIUM PHOSPHATE / pH: 7.0 / Pressure: 1 atm / Temperature: 298 K |
Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer | Type: Bruker DMX750 / Manufacturer: Bruker / Model : DMX750 / Field strength: 750 MHz : DMX750 / Field strength: 750 MHz |
|---|
- Processing
Processing
| NMR software |
| ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method: torsion angle dynamics / Software ordinal: 1 Details: REFINEMENT DETAILS ARE IN THE SUBMITTED FILE AND IN THE JRNL CITATION ABOVE. THE RESTRAINTS ARE IN THE FILES WITH THE 19 CONFORMERS. | ||||||||||||||||
| NMR ensemble | Conformer selection criteria: LOWEST TARGET FUNCTION / Conformers calculated total number: 40 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller












 PDBj
PDBj

