[English] 日本語
 Yorodumi
Yorodumi- PDB-1azm: DRUG-PROTEIN INTERACTIONS: STRUCTURE OF SULFONAMIDE DRUG COMPLEXE... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1azm | ||||||
|---|---|---|---|---|---|---|---|
| Title | DRUG-PROTEIN INTERACTIONS: STRUCTURE OF SULFONAMIDE DRUG COMPLEXED WITH HUMAN CARBONIC ANHYDRASE I | ||||||
 Components Components | CARBONIC ANHYDRASE I Carbonic anhydrase Carbonic anhydrase | ||||||
 Keywords Keywords | LYASE(OXO-ACID) / PROTEIN-DRUG INTERACTIONS / OXO-ACID LYASE /  SULFONAMIDES SULFONAMIDES | ||||||
| Function / homology |  Function and homology information Function and homology information hydro-lyase activity / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / hydro-lyase activity / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation /  cyanamide hydratase / cyanamide hydratase /  cyanamide hydratase activity / cyanamide hydratase activity /  arylesterase activity / Reversible hydration of carbon dioxide / arylesterase activity / Reversible hydration of carbon dioxide /  carbonic anhydrase / carbonic anhydrase /  carbonate dehydratase activity / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen ... carbonate dehydratase activity / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen ... hydro-lyase activity / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation / hydro-lyase activity / Gene and protein expression by JAK-STAT signaling after Interleukin-12 stimulation /  cyanamide hydratase / cyanamide hydratase /  cyanamide hydratase activity / cyanamide hydratase activity /  arylesterase activity / Reversible hydration of carbon dioxide / arylesterase activity / Reversible hydration of carbon dioxide /  carbonic anhydrase / carbonic anhydrase /  carbonate dehydratase activity / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / one-carbon metabolic process / extracellular exosome / zinc ion binding / carbonate dehydratase activity / Erythrocytes take up oxygen and release carbon dioxide / Erythrocytes take up carbon dioxide and release oxygen / one-carbon metabolic process / extracellular exosome / zinc ion binding /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  X-RAY DIFFRACTION / Resolution: 2 Å X-RAY DIFFRACTION / Resolution: 2 Å | ||||||
 Authors Authors | Chakravarty, S. / Kannan, K.K. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1994 Journal: J.Mol.Biol. / Year: 1994Title: Drug-protein interactions. Refined structures of three sulfonamide drug complexes of human carbonic anhydrase I enzyme. Authors: Chakravarty, S. / Kannan, K.K. #1:  Journal: J.Biosci. / Year: 1985 Journal: J.Biosci. / Year: 1985Title: Drug Protein Interaction at the Molecular Level: A Study of Sulphonamide Carbonic Anhydrase Complexes Authors: Chakravarty, S. / Yadava, V.S. / Kumar, V. / Kannan, K.K. #2:  Journal: Drug Action at the Molecular Level / Year: 1977 Journal: Drug Action at the Molecular Level / Year: 1977Title: Structure and Function of Carbonic Anhydrase: Comparative Studies of Sulphonamide Binding to Human Erythrocyte Carbonic Anhydrases B and C Authors: Kannan, K.K. / Vaara, I. / Notstrand, B. / Lovgren, S. / Borell, A. / Fridborg, K. / Petef, M. | ||||||
| History |
| ||||||
| Remark 700 | SHEET SHEET B1 OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE SHEET ...SHEET SHEET B1 OF THIS MOLECULE IS BIFURCATED. IN ORDER TO REPRESENT THIS FEATURE IN THE SHEET RECORDS BELOW, TWO SHEETS *B1A* AND *B1B* ARE DEFINED. STRANDS 5, 6, 7, 8, 9, AND 10 OF B1A ARE IDENTICAL TO STRANDS 2, 3, 4, 5, 6, AND 7 OF B1B, RESPECTIVELY. |
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1azm.cif.gz 1azm.cif.gz | 67.9 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1azm.ent.gz pdb1azm.ent.gz | 50.1 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1azm.json.gz 1azm.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/az/1azm https://data.pdbj.org/pub/pdb/validation_reports/az/1azm ftp://data.pdbj.org/pub/pdb/validation_reports/az/1azm ftp://data.pdbj.org/pub/pdb/validation_reports/az/1azm | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
| ||||||||
| Atom site foot note | 1: CIS PROLINE - PRO 30 / 2: CIS PROLINE - PRO 202 3: THE SULFONAMIDE DRUG 5-ACETAMIDO-1,3, 4-THIADIAZOLE-2-SULFONAMIDE HAS BEEN ASSIGNED THE THREE LETTER CODE AZM IN THE COORDINATE FILE. 4: THE DRUG MOLECULE HAS WELL DEFINED ELECTRON DENSITY IN THE ACTIVE SITE OF THE ENZYME. WHEN THE DRUG BINDS TO THE ENZYME, THE LOOP REGION COMPRISING OF RESIDUES LEU 198, THR 199 AND HIS 200 ...4: THE DRUG MOLECULE HAS WELL DEFINED ELECTRON DENSITY IN THE ACTIVE SITE OF THE ENZYME. WHEN THE DRUG BINDS TO THE ENZYME, THE LOOP REGION COMPRISING OF RESIDUES LEU 198, THR 199 AND HIS 200 UNDERGOES A SIGNIFICANT CHANGE AS COMPARED TO THE NATIVE STRUCTURE. 5: ACTIVE SITE HYDROGEN BONDED SOLVENT NETWORK INVOLVING HIS 67 AND HIS 200 IS POSSIBLY IMPORTANT FOR THE CATALYTIC ACTIVITY AND INHIBITION OF THIS ISOENZYME. 6: ZINC ZN(II) IS THE CATALYTICALLY ESSENTIALL ZINC ION. |
- Components
Components
| #1: Protein |  Carbonic anhydrase Carbonic anhydraseMass: 28774.988 Da / Num. of mol.: 1 Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / References: UniProt: P00915, Homo sapiens (human) / References: UniProt: P00915,  carbonic anhydrase carbonic anhydrase |
|---|---|
| #2: Chemical | ChemComp-ZN / |
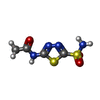
| #3: Chemical | ChemComp-AZM /  Acetazolamide Acetazolamide |
| #4: Water | ChemComp-HOH /  Water Water |
| Compound details | THE DRUG MOLECULE HAS WELL DEFINED ELECTRON DENSITY IN THE ACTIVE SITE OF THE ENZYME. WHEN THE DRUG ...THE DRUG MOLECULE HAS WELL DEFINED ELECTRON DENSITY IN THE ACTIVE SITE OF THE ENZYME. WHEN THE DRUG BINDS TO THE ENZYME, THE LOOP REGION COMPRISING |
| Nonpolymer details | ZINC ZN(II) IS THE CATALYTICA |
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION X-RAY DIFFRACTION |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 2.02 Å3/Da / Density % sol: 39.14 % |
|---|---|
Crystal grow | *PLUS Method: unknown |
- Processing
Processing
| Software | Name: PROTEIN / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Resolution: 2→10 Å /
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2→10 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.195 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS |
 Movie
Movie Controller
Controller
























 PDBj
PDBj