+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1aoj | ||||||
|---|---|---|---|---|---|---|---|
| Title | THE SH3 DOMAIN OF EPS8 EXISTS AS A NOVEL INTERTWINED DIMER | ||||||
 Components Components | EPS8 | ||||||
 Keywords Keywords |  SIGNAL TRANSDUCTION / SIGNAL TRANSDUCTION /  SH3 DOMAIN / SH3 DOMAIN /  EPS8 / PROLINE RICH PEPTIDE EPS8 / PROLINE RICH PEPTIDE | ||||||
| Function / homology |  Function and homology information Function and homology informationregulation of actin filament length / actin polymerization-dependent cell motility / dendritic cell migration / stereocilium tip / actin crosslink formation / stereocilium bundle / behavioral response to ethanol / stereocilium / exit from mitosis / regulation of Rho protein signal transduction ...regulation of actin filament length / actin polymerization-dependent cell motility / dendritic cell migration / stereocilium tip / actin crosslink formation / stereocilium bundle / behavioral response to ethanol / stereocilium / exit from mitosis / regulation of Rho protein signal transduction / barbed-end actin filament capping / NMDA selective glutamate receptor complex / positive regulation of ruffle assembly / Rac protein signal transduction / regulation of postsynaptic membrane neurotransmitter receptor levels / actin filament bundle assembly /  brush border / Rho protein signal transduction / cellular response to leukemia inhibitory factor / adult locomotory behavior / ruffle membrane / brush border / Rho protein signal transduction / cellular response to leukemia inhibitory factor / adult locomotory behavior / ruffle membrane /  small GTPase binding / small GTPase binding /  actin binding / actin binding /  cell cortex / regulation of cell shape / cell cortex / regulation of cell shape /  growth cone / actin cytoskeleton organization / growth cone / actin cytoskeleton organization /  postsynaptic density / glutamatergic synapse / postsynaptic density / glutamatergic synapse /  synapse / synapse /  plasma membrane / plasma membrane /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Mus musculus (house mouse) Mus musculus (house mouse) | ||||||
| Method |  X-RAY DIFFRACTION / X-RAY DIFFRACTION /  MOLECULAR REPLACEMENT / Resolution: 2.5 Å MOLECULAR REPLACEMENT / Resolution: 2.5 Å | ||||||
 Authors Authors | Kishan, K.V.R. / Newcomer, M.E. | ||||||
 Citation Citation |  Journal: Nat.Struct.Biol. / Year: 1997 Journal: Nat.Struct.Biol. / Year: 1997Title: The SH3 domain of Eps8 exists as a novel intertwined dimer. Authors: Kishan, K.V. / Scita, G. / Wong, W.T. / Di Fiore, P.P. / Newcomer, M.E. #1:  Journal: Embo J. / Year: 1993 Journal: Embo J. / Year: 1993Title: Eps8, a Substrate for the Epidermal Growth Factor Receptor Kinase, Enhances Egf-Dependent Mitogenic Signals Authors: Fazioli, F. / Minichiello, L. / Matoska, V. / Castagnino, P. / Miki, T. / Wong, W.T. / Di Fiore, P.P. | ||||||
| History |
|
- Structure visualization
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1aoj.cif.gz 1aoj.cif.gz | 33.3 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1aoj.ent.gz pdb1aoj.ent.gz | 24.8 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1aoj.json.gz 1aoj.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ao/1aoj https://data.pdbj.org/pub/pdb/validation_reports/ao/1aoj ftp://data.pdbj.org/pub/pdb/validation_reports/ao/1aoj ftp://data.pdbj.org/pub/pdb/validation_reports/ao/1aoj | HTTPS FTP |
|---|
-Related structure data
| Related structure data |  1shfS S: Starting model for refinement |
|---|---|
| Similar structure data |
- Links
Links
- Assembly
Assembly
| Deposited unit | 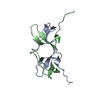
| ||||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 |
| ||||||||
| Unit cell |
|
- Components
Components
| #1: Protein |  / SRC HOMOLOGY DOMAIN / SRC HOMOLOGY DOMAINMass: 7044.960 Da / Num. of mol.: 2 / Fragment: SH3 DOMAIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Mus musculus (house mouse) / Production host: Mus musculus (house mouse) / Production host:   Escherichia coli (E. coli) / References: UniProt: Q08509 Escherichia coli (E. coli) / References: UniProt: Q08509#2: Water | ChemComp-HOH / |  Water Water |
|---|
-Experimental details
-Experiment
| Experiment | Method:  X-RAY DIFFRACTION / Number of used crystals: 2 X-RAY DIFFRACTION / Number of used crystals: 2 |
|---|
- Sample preparation
Sample preparation
| Crystal | Density Matthews: 1.85 Å3/Da / Density % sol: 33.52 % | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Crystal grow | pH: 7.5 Details: PROTEIN WAS CRYSTALLIZED FROM 0.4M SODIUM POTASSIUM TARTRATE, PH 7.5. PROTEIN CONC. 6MG/ML. CRYSTALLIZATION TIME 2-3 WEEKS. | |||||||||||||||
| Crystal grow | *PLUS pH: 7.4 / Method: vapor diffusion, hanging drop | |||||||||||||||
| Components of the solutions | *PLUS
|
-Data collection
| Diffraction | Mean temperature: 300 K |
|---|---|
| Diffraction source | Source:  ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 ROTATING ANODE / Type: RIGAKU RUH2R / Wavelength: 1.5418 |
| Detector | Type: RIGAKU RAXIS IIC / Detector: IMAGE PLATE / Date: Feb 1, 1996 / Details: MSC/YALE MIRRORS |
| Radiation | Monochromatic (M) / Laue (L): M / Scattering type: x-ray |
| Radiation wavelength | Wavelength : 1.5418 Å / Relative weight: 1 : 1.5418 Å / Relative weight: 1 |
| Reflection | Resolution: 2.5→32 Å / Num. obs: 3406 / % possible obs: 97 % / Observed criterion σ(I): 4 / Redundancy: 6.2 % / Rmerge(I) obs: 0.106 / Net I/σ(I): 12.6 |
| Reflection shell | Resolution: 2.5→2.6 Å / Redundancy: 1.8 % / Rmerge(I) obs: 0.397 / Mean I/σ(I) obs: 3.3 / % possible all: 92 |
| Reflection | *PLUS Num. measured all: 21281 |
| Reflection shell | *PLUS % possible obs: 92 % |
- Processing
Processing
| Software |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Refinement | Method to determine structure : :  MOLECULAR REPLACEMENT MOLECULAR REPLACEMENTStarting model: PDB ENTRY 1SHF Resolution: 2.5→32 Å / Isotropic thermal model: TNT BCORREL / Cross valid method: R-FREE THROUGHOUT / σ(F): 2 / Stereochemistry target values: TNT PROTGEO Details: REFINEMENT FIRST STARTED WITH X-PLOR SIMULATED ANNEALING PROTOCOL
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | Bsol: 192 Å2 / ksol: 0.679 e/Å3 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement step | Cycle: LAST / Resolution: 2.5→32 Å
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints |
| ||||||||||||||||||||||||||||||||||||||||||||||||||
| Software | *PLUS Name: TNT / Version: 5E / Classification: refinement | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refinement | *PLUS Rfactor obs: 0.193 / Rfactor Rfree : 0.277 : 0.277 | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Solvent computation | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Displacement parameters | *PLUS | ||||||||||||||||||||||||||||||||||||||||||||||||||
| Refine LS restraints | *PLUS
|
 Movie
Movie Controller
Controller











 PDBj
PDBj

