+ Open data
Open data
- Basic information
Basic information
| Entry | Database: PDB / ID: 1neb | ||||||
|---|---|---|---|---|---|---|---|
| Title | SH3 DOMAIN FROM HUMAN NEBULIN, NMR, MINIMIZED AVERAGE STRUCTURE | ||||||
 Components Components | NEBULIN | ||||||
 Keywords Keywords |  SH3 DOMAIN / SH3 DOMAIN /  NEBULIN / NEBULIN /  Z-DISK ASSEMBLY / ACTIN-BINDING Z-DISK ASSEMBLY / ACTIN-BINDING | ||||||
| Function / homology |  Function and homology information Function and homology informationcardiac muscle thin filament assembly / regulation of actin filament length / somatic muscle development / Striated Muscle Contraction / muscle organ development / structural constituent of muscle / Z disc /  actin filament binding / actin filament binding /  actin cytoskeleton / extracellular exosome / actin cytoskeleton / extracellular exosome /  cytosol cytosolSimilarity search - Function | ||||||
| Biological species |   Homo sapiens (human) Homo sapiens (human) | ||||||
| Method |  SOLUTION NMR / DISTANCE GEOMETRY, SOLUTION NMR / DISTANCE GEOMETRY,  SIMULATED ANNEALING SIMULATED ANNEALING | ||||||
 Authors Authors | Politou, A.S. / Pastore, A. | ||||||
 Citation Citation |  Journal: J.Mol.Biol. / Year: 1998 Journal: J.Mol.Biol. / Year: 1998Title: SH3 in muscles: solution structure of the SH3 domain from nebulin. Authors: Politou, A.S. / Millevoi, S. / Gautel, M. / Kolmerer, B. / Pastore, A. | ||||||
| History |
|
- Structure visualization
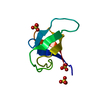
Structure visualization
| Structure viewer | Molecule:  Molmil Molmil Jmol/JSmol Jmol/JSmol |
|---|
- Downloads & links
Downloads & links
- Download
Download
| PDBx/mmCIF format |  1neb.cif.gz 1neb.cif.gz | 26.4 KB | Display |  PDBx/mmCIF format PDBx/mmCIF format |
|---|---|---|---|---|
| PDB format |  pdb1neb.ent.gz pdb1neb.ent.gz | 20 KB | Display |  PDB format PDB format |
| PDBx/mmJSON format |  1neb.json.gz 1neb.json.gz | Tree view |  PDBx/mmJSON format PDBx/mmJSON format | |
| Others |  Other downloads Other downloads |
-Validation report
| Arichive directory |  https://data.pdbj.org/pub/pdb/validation_reports/ne/1neb https://data.pdbj.org/pub/pdb/validation_reports/ne/1neb ftp://data.pdbj.org/pub/pdb/validation_reports/ne/1neb ftp://data.pdbj.org/pub/pdb/validation_reports/ne/1neb | HTTPS FTP |
|---|
-Related structure data
- Links
Links
- Assembly
Assembly
| Deposited unit | 
| |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 |
| |||||||||
| NMR ensembles |
|
- Components
Components
| #1: Protein |  Mass: 6614.411 Da / Num. of mol.: 1 / Fragment: SH3 DOMAIN Source method: isolated from a genetically manipulated source Source: (gene. exp.)   Homo sapiens (human) / Cell line: BL21 / Plasmid: BL21 / Production host: Homo sapiens (human) / Cell line: BL21 / Plasmid: BL21 / Production host:   Escherichia coli (E. coli) / Strain (production host): BL21 (DE3)-[PLYSS] / References: UniProt: P20929 Escherichia coli (E. coli) / Strain (production host): BL21 (DE3)-[PLYSS] / References: UniProt: P20929 |
|---|
-Experimental details
-Experiment
| Experiment | Method:  SOLUTION NMR SOLUTION NMR | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR experiment |
|
- Sample preparation
Sample preparation
| Sample conditions | pH: 6.9 / Temperature: 300 K |
|---|---|
Crystal grow | *PLUS Method: other / Details: NMR |
-NMR measurement
| NMR spectrometer |
|
|---|
- Processing
Processing
| Software |
| ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NMR software |
| ||||||||||||
| Refinement | Method: DISTANCE GEOMETRY,  SIMULATED ANNEALING / Software ordinal: 1 / Details: REFINEMENT DETAILS ARE IN THE SUBMITTED PAPER. SIMULATED ANNEALING / Software ordinal: 1 / Details: REFINEMENT DETAILS ARE IN THE SUBMITTED PAPER. | ||||||||||||
| NMR ensemble | Conformer selection criteria: LEAST RESTRAINT VIOLATION / Conformers calculated total number: 100 / Conformers submitted total number: 1 |
 Movie
Movie Controller
Controller












 PDBj
PDBj

