+ データを開く
データを開く
- 基本情報
基本情報
| 登録情報 | データベース: EMDB / ID: EMD-7076 | |||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| タイトル | Mechanisms of Opening and Closing of the Bacterial Replicative Helicase: The DnaB Helicase and Lambda P Helicase Loader Complex | |||||||||||||||||||||||||||||||||||||||
 マップデータ マップデータ | DnaB-LambdaP helicase-helicase loader complex from single particle cryoEM at 4.1A. The suggested viewing thresholds are 0.0276 (Chimera) or an isomesh level of 6 (PyMol). | |||||||||||||||||||||||||||||||||||||||
 試料 試料 |
| |||||||||||||||||||||||||||||||||||||||
 キーワード キーワード | Helicase Loader /  Helicase (ヘリカーゼ) / Helicase (ヘリカーゼ) /  DNA replication (DNA複製) / DNA replication (DNA複製) /  ATPase (ATPアーゼ) / DNA Replication Initiation / ATPase (ATPアーゼ) / DNA Replication Initiation /  Bacteriophage Lambda / Bacteriophage Lambda /  REPLICATION (DNA複製) REPLICATION (DNA複製) | |||||||||||||||||||||||||||||||||||||||
| 機能・相同性 |  機能・相同性情報 機能・相同性情報DnaB-DnaC complex / DnaB-DnaC-Rep-PriC complex / DnaB-DnaG complex / DnaB-DnaC-DnaT-PriA-PriC complex /  DNA helicase complex / DnaB-DnaC-DnaT-PriA-PriB complex / DNA helicase complex / DnaB-DnaC-DnaT-PriA-PriB complex /  primosome complex / bidirectional double-stranded viral DNA replication / primosome complex / bidirectional double-stranded viral DNA replication /  replisome / replisome /  DNA replication, synthesis of primer ...DnaB-DnaC complex / DnaB-DnaC-Rep-PriC complex / DnaB-DnaG complex / DnaB-DnaC-DnaT-PriA-PriC complex / DNA replication, synthesis of primer ...DnaB-DnaC complex / DnaB-DnaC-Rep-PriC complex / DnaB-DnaG complex / DnaB-DnaC-DnaT-PriA-PriC complex /  DNA helicase complex / DnaB-DnaC-DnaT-PriA-PriB complex / DNA helicase complex / DnaB-DnaC-DnaT-PriA-PriB complex /  primosome complex / bidirectional double-stranded viral DNA replication / primosome complex / bidirectional double-stranded viral DNA replication /  replisome / replisome /  DNA replication, synthesis of primer / DNA duplex unwinding / response to ionizing radiation / replication fork processing / DNA unwinding involved in DNA replication / DNA replication initiation / DNA replication, synthesis of primer / DNA duplex unwinding / response to ionizing radiation / replication fork processing / DNA unwinding involved in DNA replication / DNA replication initiation /  DNA helicase activity / DNA helicase activity /  helicase activity / helicase activity /  ヘリカーゼ / ヘリカーゼ /  DNA複製 / DNA複製 /  hydrolase activity / hydrolase activity /  ATP hydrolysis activity / ATP hydrolysis activity /  DNA binding / DNA binding /  ATP binding / identical protein binding / ATP binding / identical protein binding /  細胞質基質 細胞質基質類似検索 - 分子機能 | |||||||||||||||||||||||||||||||||||||||
| 生物種 |   Escherichia coli (大腸菌) / Escherichia coli (大腸菌) /   Enterobacteria phage lambda (λファージ) / Enterobacteria phage lambda (λファージ) /   Escherichia coli O111:NM (大腸菌) / Escherichia coli O111:NM (大腸菌) /   Escherichia phage lambda (λファージ) Escherichia phage lambda (λファージ) | |||||||||||||||||||||||||||||||||||||||
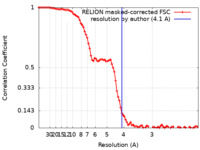
| 手法 |  単粒子再構成法 / 単粒子再構成法 /  クライオ電子顕微鏡法 / 解像度: 4.1 Å クライオ電子顕微鏡法 / 解像度: 4.1 Å | |||||||||||||||||||||||||||||||||||||||
 データ登録者 データ登録者 | Chase J / Catalano A | |||||||||||||||||||||||||||||||||||||||
| 資金援助 |  米国, 12件 米国, 12件
| |||||||||||||||||||||||||||||||||||||||
 引用 引用 |  ジャーナル: Elife / 年: 2018 ジャーナル: Elife / 年: 2018タイトル: Mechanisms of opening and closing of the bacterial replicative helicase. 著者: Jillian Chase / Andrew Catalano / Alex J Noble / Edward T Eng / Paul Db Olinares / Kelly Molloy / Danaya Pakotiprapha / Martin Samuels / Brian Chait / Amedee des Georges / David Jeruzalmi /   要旨: Assembly of bacterial ring-shaped hexameric replicative helicases on single-stranded (ss) DNA requires specialized loading factors. However, mechanisms implemented by these factors during opening and ...Assembly of bacterial ring-shaped hexameric replicative helicases on single-stranded (ss) DNA requires specialized loading factors. However, mechanisms implemented by these factors during opening and closing of the helicase, which enable and restrict access to an internal chamber, are not known. Here, we investigate these mechanisms in the DnaB helicase•bacteriophage λ helicase loader (λP) complex. We show that five copies of λP bind at DnaB subunit interfaces and reconfigure the helicase into an open spiral conformation that is intermediate to previously observed closed ring and closed spiral forms; reconfiguration also produces openings large enough to admit ssDNA into the inner chamber. The helicase is also observed in a restrained inactive configuration that poises it to close on activating signal, and transition to the translocation state. Our findings provide insights into helicase opening, delivery to the origin and ssDNA entry, and closing in preparation for translocation. | |||||||||||||||||||||||||||||||||||||||
| 履歴 |
|
- 構造の表示
構造の表示
| ムービー |
 ムービービューア ムービービューア |
|---|---|
| 構造ビューア | EMマップ:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| 添付画像 |
- ダウンロードとリンク
ダウンロードとリンク
-EMDBアーカイブ
| マップデータ |  emd_7076.map.gz emd_7076.map.gz | 59.9 MB |  EMDBマップデータ形式 EMDBマップデータ形式 | |
|---|---|---|---|---|
| ヘッダ (付随情報) |  emd-7076-v30.xml emd-7076-v30.xml emd-7076.xml emd-7076.xml | 26.1 KB 26.1 KB | 表示 表示 |  EMDBヘッダ EMDBヘッダ |
| FSC (解像度算出) |  emd_7076_fsc.xml emd_7076_fsc.xml | 9.2 KB | 表示 |  FSCデータファイル FSCデータファイル |
| 画像 |  emd_7076.png emd_7076.png | 157.3 KB | ||
| Filedesc metadata |  emd-7076.cif.gz emd-7076.cif.gz | 9.8 KB | ||
| アーカイブディレクトリ |  http://ftp.pdbj.org/pub/emdb/structures/EMD-7076 http://ftp.pdbj.org/pub/emdb/structures/EMD-7076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7076 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-7076 | HTTPS FTP |
-関連構造データ
- リンク
リンク
| EMDBのページ |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| 「今月の分子」の関連する項目 |
- マップ
マップ
| ファイル |  ダウンロード / ファイル: emd_7076.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) ダウンロード / ファイル: emd_7076.map.gz / 形式: CCP4 / 大きさ: 64 MB / タイプ: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 注釈 | DnaB-LambdaP helicase-helicase loader complex from single particle cryoEM at 4.1A. The suggested viewing thresholds are 0.0276 (Chimera) or an isomesh level of 6 (PyMol). | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ボクセルのサイズ | X=Y=Z: 1.07 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 密度 |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 対称性 | 空間群: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 詳細 | EMDB XML:
CCP4マップ ヘッダ情報:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-添付データ
- 試料の構成要素
試料の構成要素
-全体 : DnaB helicase - Lambda P helicase loader DNA replication complex
| 全体 | 名称: DnaB helicase - Lambda P helicase loader DNA replication complex |
|---|---|
| 要素 |
|
-超分子 #1: DnaB helicase - Lambda P helicase loader DNA replication complex
| 超分子 | 名称: DnaB helicase - Lambda P helicase loader DNA replication complex タイプ: complex / ID: 1 / 親要素: 0 / 含まれる分子: #1-#2 詳細: pET24a containing full-length DnaB was co-expressed with pCDFDuet containing full-length LambdaP in BL21(DE3) cells. The resolution of the LambdaP portion of our EM map did not permit the ...詳細: pET24a containing full-length DnaB was co-expressed with pCDFDuet containing full-length LambdaP in BL21(DE3) cells. The resolution of the LambdaP portion of our EM map did not permit the unambiguous assignment of the amino acid sequence to the structure. As such, the model for LambdaP was built as a poly alanine model. Additionally, only half of LambdaP was observed in our maps due to the intrinsic flexibility of the amino and carboxy terminal domains of LambdaP. Subsequent experiments determined that the observed portion of LambdaP in our maps corresponds to the C-terminal domain. |
|---|---|
| 由来(天然) | 生物種:   Escherichia coli (大腸菌) Escherichia coli (大腸菌) |
-超分子 #2: E coli DnaB helicase
| 超分子 | 名称: E coli DnaB helicase / タイプ: complex / ID: 2 / 親要素: 1 / 含まれる分子: #1 詳細: E coli DnaB helicase is observed as an open-spiral hexamer, in which one of the interfaces is breached. Five ADP molecules are observed at the five intact ATP binding sites. Additionally, ...詳細: E coli DnaB helicase is observed as an open-spiral hexamer, in which one of the interfaces is breached. Five ADP molecules are observed at the five intact ATP binding sites. Additionally, clear density is observed for five of six linkers permitting unambiguous assignment of NTD to parent CTD domain. |
|---|---|
| 由来(天然) | 生物種:   Escherichia coli (大腸菌) Escherichia coli (大腸菌) |
-超分子 #3: Lambda P helicase loader
| 超分子 | 名称: Lambda P helicase loader / タイプ: complex / ID: 3 / 親要素: 1 / 含まれる分子: #2 詳細: Five lambda P molecules were observed bound to the five intact DnaB subunit interfaces. Unambiguous assignment of side chain density for lambda P was not possible due to the resolution of ...詳細: Five lambda P molecules were observed bound to the five intact DnaB subunit interfaces. Unambiguous assignment of side chain density for lambda P was not possible due to the resolution of this region of the EM map. Instead, a polyalanine model was built for each lambda P molecule. Additionally, density for approximately half of the expected 233 residues of lambda P was observed owing to flexibility between domains. Subsequent experiments confirmed that the observed region of Lambda P is the C-terminal domain, which interacts with DnaB. |
|---|---|
| 由来(天然) | 生物種:   Enterobacteria phage lambda (λファージ) Enterobacteria phage lambda (λファージ) |
-分子 #1: Replicative DNA helicase
| 分子 | 名称: Replicative DNA helicase / タイプ: protein_or_peptide / ID: 1 / コピー数: 6 / 光学異性体: LEVO / EC番号:  ヘリカーゼ ヘリカーゼ |
|---|---|
| 由来(天然) | 生物種:   Escherichia coli O111:NM (大腸菌) Escherichia coli O111:NM (大腸菌) |
| 分子量 | 理論値: 52.450945 KDa |
| 組換発現 | 生物種:   Escherichia coli BL21(DE3) (大腸菌) Escherichia coli BL21(DE3) (大腸菌) |
| 配列 | 文字列: MAGNKPFNKQ QAEPRERDPQ VAGLKVPPHS IEAEQSVLGG LMLDNERWDD VAERVVADDF YTRPHRHIFT EMARLQESGS PIDLITLAE SLERQGQLDS VGGFAYLAEL SKNTPSAANI SAYADIVRER AVVREMISVA NEIAEAGFDP QGRTSEDLLD L AESRVFKI ...文字列: MAGNKPFNKQ QAEPRERDPQ VAGLKVPPHS IEAEQSVLGG LMLDNERWDD VAERVVADDF YTRPHRHIFT EMARLQESGS PIDLITLAE SLERQGQLDS VGGFAYLAEL SKNTPSAANI SAYADIVRER AVVREMISVA NEIAEAGFDP QGRTSEDLLD L AESRVFKI AESRANKDEG PKNIADVLDA TVARIEQLFQ QPHDGVTGVN TGYDDLNKKT AGLQPSDLII VAARPSMGKT TF AMNLVEN AAMLQDKPVL IFSLEMPSEQ IMMRSLASLS RVDQTKIRTG QLDDEDWARI SGTMGILLEK RNIYIDDSSG LTP TEVRSR ARRIAREHGG IGLIMIDYLQ LMRVPALSDN RTLEIAEISR SLKALAKELN VPVVALSQLN RSLEQRADKR PVNS DLRES GSIEQDADLI MFIYRDEVYH ENSDLKGIAE IIIGKQRNGP IGTVRLTFNG QWSRFDNYAG PQYDDE UniProtKB: Replicative DNA helicase |
-分子 #2: Replication protein P
| 分子 | 名称: Replication protein P / タイプ: protein_or_peptide / ID: 2 / コピー数: 5 / 光学異性体: LEVO |
|---|---|
| 由来(天然) | 生物種:   Escherichia phage lambda (λファージ) Escherichia phage lambda (λファージ) |
| 分子量 | 理論値: 23.141221 KDa |
| 組換発現 | 生物種:   Escherichia coli BL21(DE3) (大腸菌) Escherichia coli BL21(DE3) (大腸菌) |
| 配列 | 文字列: MKNIAAQMVN FDREQMRRIA NNMPEQYDEK PQVQQVAQII NGVFSQLLAT FPASLANRDQ NEVNEIRRQW VLAFRENGIT TMEQVNAGM RVARRQNRPF LPSPGQFV(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK) ...文字列: MKNIAAQMVN FDREQMRRIA NNMPEQYDEK PQVQQVAQII NGVFSQLLAT FPASLANRDQ NEVNEIRRQW VLAFRENGIT TMEQVNAGM RVARRQNRPF LPSPGQFV(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK)(UNK)(UNK)(UNK)(UNK)(UNK) (UNK)(UNK) (UNK)(UNK) UniProtKB:  Replication protein P Replication protein P |
-分子 #3: ADENOSINE-5'-DIPHOSPHATE
| 分子 | 名称: ADENOSINE-5'-DIPHOSPHATE / タイプ: ligand / ID: 3 / コピー数: 5 / 式: ADP |
|---|---|
| 分子量 | 理論値: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-実験情報
-構造解析
| 手法 |  クライオ電子顕微鏡法 クライオ電子顕微鏡法 |
|---|---|
 解析 解析 |  単粒子再構成法 単粒子再構成法 |
| 試料の集合状態 | particle |
- 試料調製
試料調製
| 濃度 | 0.48 mg/mL | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 緩衝液 | pH: 7.5 構成要素:
詳細: Concentrated BP sample (18mg/mL) was diluted with freshly prepared buffer to desired concentration (~1.5 micromolar) for grid preparation. | ||||||||||||||||||
| グリッド | モデル: Quantifoil R0.6/1 / 材質: GOLD / メッシュ: 400 / 支持フィルム - 材質: CARBON / 支持フィルム - トポロジー: CONTINUOUS / 前処理 - タイプ: PLASMA CLEANING / 前処理 - 時間: 60 sec. / 前処理 - 雰囲気: OTHER 詳細: The grid was coated with 50 nm of evaporated gold prior to use. All remaining carbon was removed by plasma cleaning for 5 minutes in a Gatan Solarus plasma cleaner. | ||||||||||||||||||
| 凍結 | 凍結剤: ETHANE / チャンバー内湿度: 100 % / チャンバー内温度: 277.15 K / 装置: FEI VITROBOT MARK IV 詳細: 3uL of sample was adhered to a fresh plasma cleaned grid and allowed to adsorb for 30 seconds, blotted for 3 seconds with a blot force of 4 and plunge frozen into liquid nitrogen-cooled ethane.. | ||||||||||||||||||
| 詳細 | The sample was monodisperse. Side views were more electron weak than top or bottom views creating challenges for particle picking. This issue was overcome with cryo-electron tomography techniques used for 1) initial model generation and 2) template generation for particle picking. |
- 電子顕微鏡法
電子顕微鏡法
| 顕微鏡 | FEI TITAN KRIOS |
|---|---|
| 電子線 | 加速電圧: 300 kV / 電子線源:  FIELD EMISSION GUN FIELD EMISSION GUN |
| 電子光学系 | C2レンズ絞り径: 70.0 µm / 照射モード: FLOOD BEAM / 撮影モード: BRIGHT FIELD Bright-field microscopy / Cs: 2.7 mm / 最大 デフォーカス(公称値): -0.003 µm / 最小 デフォーカス(公称値): -0.001 µm / 倍率(公称値): 22500 Bright-field microscopy / Cs: 2.7 mm / 最大 デフォーカス(公称値): -0.003 µm / 最小 デフォーカス(公称値): -0.001 µm / 倍率(公称値): 22500 |
| 特殊光学系 | 球面収差補正装置: The Krios this data was collected on has a Cs of 2.7. |
| 試料ステージ | 試料ホルダーモデル: FEI TITAN KRIOS AUTOGRID HOLDER ホルダー冷却材: NITROGEN |
| 温度 | 最低: 70.0 K / 最高: 70.0 K |
| 詳細 | Preliminary grid screening was performed prior to Krios data collections. All microscope alignments were completed by the New York Structural Biology SEMC team. |
| 撮影 | フィルム・検出器のモデル: GATAN K2 SUMMIT (4k x 4k) 検出モード: COUNTING / デジタル化 - サイズ - 横: 3838 pixel / デジタル化 - サイズ - 縦: 3710 pixel / デジタル化 - 画像ごとのフレーム数: 1-50 / 撮影したグリッド数: 3 / 実像数: 2426 / 平均露光時間: 10.0 sec. / 平均電子線量: 8.0 e/Å2 詳細: Single particle movies were recorded at a pixel size of 1.07 angstroms/pixel. Three 24-hour sessions produced 2,426 micrograph movies. In addition, five tilt series were collected from the ...詳細: Single particle movies were recorded at a pixel size of 1.07 angstroms/pixel. Three 24-hour sessions produced 2,426 micrograph movies. In addition, five tilt series were collected from the same grids bi-directionally over a tilt range of -45 degrees to +45 degrees in 3 degree increments at a dose of 2.57 to 3.3 electrons per angstrom squared (total accumulated dose of 90 electrons per angstrom squared). Tilt series were collected at a pixel size of 1.76 angstroms and at defocus values of -2.8um, -6.1um and -9.3um. |
| 実験機器 |  モデル: Titan Krios / 画像提供: FEI Company |
- 画像解析
画像解析
-原子モデル構築 1
| 初期モデル |
| ||||||
|---|---|---|---|---|---|---|---|
| 詳細 | The initial fitting was done with the 2R5U and 3BH0 models onto which the E. coli amino sequence had been built. The linker segments that connected these segments were built by hand. PHENIX real_space_refine was used to refine the complete model for the B6P5 entity. | ||||||
| 精密化 | 空間: REAL / プロトコル: FLEXIBLE FIT | ||||||
| 得られたモデル |  PDB-6bbm: |
 ムービー
ムービー コントローラー
コントローラー