[English] 日本語
 Yorodumi
Yorodumi- EMDB-24489: The C1b projection within the ciliary central apparatus from the ... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-24489 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | The C1b projection within the ciliary central apparatus from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas fap413 mutant axonemes | |||||||||
 Map data Map data | fap413 mutant C1b projection | |||||||||
 Sample Sample |
| |||||||||
| Biological species |  | |||||||||
| Method | electron tomography / cryo EM / Resolution: 25.0 Å | |||||||||
 Authors Authors | Cai K / Nicastro D | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: J Cell Sci / Year: 2021 Journal: J Cell Sci / Year: 2021Title: Structural organization of the C1b projection within the ciliary central apparatus. Authors: Kai Cai / Yanhe Zhao / Lei Zhao / Nhan Phan / Yuqing Hou / Xi Cheng / George B Witman / Daniela Nicastro /  Abstract: Motile cilia have a '9+2' structure containing nine doublet microtubules and a central apparatus (CA) composed of two singlet microtubules with associated projections. The CA plays crucial roles in ...Motile cilia have a '9+2' structure containing nine doublet microtubules and a central apparatus (CA) composed of two singlet microtubules with associated projections. The CA plays crucial roles in regulating ciliary motility. Defects in CA assembly or function usually result in motility-impaired or paralyzed cilia, which in humans causes disease. Despite their importance, the protein composition and functions of most CA projections remain largely unknown. Here, we combined genetic, proteomic and cryo-electron tomographic approaches to compare the CA of wild-type Chlamydomonas reinhardtii with those of three CA mutants. Our results show that two proteins, FAP42 and FAP246, are localized to the L-shaped C1b projection of the CA, where they interact with the candidate CA protein FAP413. FAP42 is a large protein that forms the peripheral 'beam' of the C1b projection, and the FAP246-FAP413 subcomplex serves as the 'bracket' between the beam (FAP42) and the C1b 'pillar' that attaches the projection to the C1 microtubule. The FAP246-FAP413-FAP42 complex is essential for stable assembly of the C1b, C1f and C2b projections, and loss of these proteins leads to ciliary motility defects. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_24489.map.gz emd_24489.map.gz | 2.1 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-24489-v30.xml emd-24489-v30.xml emd-24489.xml emd-24489.xml | 9.7 KB 9.7 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_24489.png emd_24489.png | 108 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-24489 http://ftp.pdbj.org/pub/emdb/structures/EMD-24489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24489 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-24489 | HTTPS FTP |
-Validation report
| Summary document |  emd_24489_validation.pdf.gz emd_24489_validation.pdf.gz | 294 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_24489_full_validation.pdf.gz emd_24489_full_validation.pdf.gz | 293.6 KB | Display | |
| Data in XML |  emd_24489_validation.xml.gz emd_24489_validation.xml.gz | 4.7 KB | Display | |
| Data in CIF |  emd_24489_validation.cif.gz emd_24489_validation.cif.gz | 5.2 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24489 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24489 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24489 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-24489 | HTTPS FTP |
-Related structure data
| Related structure data | C: citing same article ( |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_24489.map.gz / Format: CCP4 / Size: 2.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_24489.map.gz / Format: CCP4 / Size: 2.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | fap413 mutant C1b projection | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
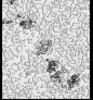
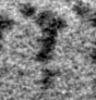
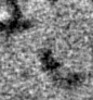
| Projections & slices | Image control
Images are generated by Spider. generated in cubic-lattice coordinate | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 3.151 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
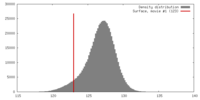
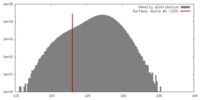
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : The C1b projection within the the ciliary central apparatus from ...
| Entire | Name: The C1b projection within the the ciliary central apparatus from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas wild type axonemes |
|---|---|
| Components |
|
-Supramolecule #1: The C1b projection within the the ciliary central apparatus from ...
| Supramolecule | Name: The C1b projection within the the ciliary central apparatus from the cryo-electron tomography and subtomographic average of isolated Chlamydomonas wild type axonemes type: organelle_or_cellular_component / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:  |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | electron tomography |
| Aggregation state | cell |
- Sample preparation
Sample preparation
| Buffer | pH: 7.4 |
|---|---|
| Grid | Model: Quantifoil R2/2 / Material: COPPER / Mesh: 200 / Support film - Material: CARBON / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
| Sectioning | Other: NO SECTIONING |
| Fiducial marker | Manufacturer: Sigma Aldrich / Diameter: 10 nm |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Specialist optics | Phase plate: VOLTA PHASE PLATE |
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Detector mode: COUNTING / Average electron dose: 1.5 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | C2 aperture diameter: 100.0 µm / Illumination mode: SPOT SCAN / Imaging mode: BRIGHT FIELD |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| Final reconstruction | Algorithm: BACK PROJECTION / Resolution.type: BY AUTHOR / Resolution: 25.0 Å / Resolution method: FSC 0.5 CUT-OFF / Software - Name: eTomo / Number images used: 1401 |
|---|
 Movie
Movie Controller
Controller










 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)