[English] 日本語
 Yorodumi
Yorodumi- EMDB-23145: Cryo-electron microscopy reconstruction of locally refined antibo... -
+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-23145 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | Cryo-electron microscopy reconstruction of locally refined antibody DH898.1 Fab-dimer | |||||||||
 Map data Map data | Final density modified, masked, locally refined map of DH898.1 Fab dimer | |||||||||
 Sample Sample |
| |||||||||
| Biological species |    Human immunodeficiency virus 1 Human immunodeficiency virus 1 | |||||||||
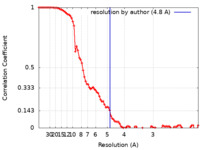
| Method | single particle reconstruction / cryo EM / Resolution: 4.8 Å | |||||||||
 Authors Authors | Edwards RJ / Acharya P | |||||||||
| Funding support |  United States, 2 items United States, 2 items
| |||||||||
 Citation Citation |  Journal: Cell / Year: 2021 Journal: Cell / Year: 2021Title: Fab-dimerized glycan-reactive antibodies are a structural category of natural antibodies. Authors: Wilton B Williams / R Ryan Meyerhoff / R J Edwards / Hui Li / Kartik Manne / Nathan I Nicely / Rory Henderson / Ye Zhou / Katarzyna Janowska / Katayoun Mansouri / Sophie Gobeil / Tyler ...Authors: Wilton B Williams / R Ryan Meyerhoff / R J Edwards / Hui Li / Kartik Manne / Nathan I Nicely / Rory Henderson / Ye Zhou / Katarzyna Janowska / Katayoun Mansouri / Sophie Gobeil / Tyler Evangelous / Bhavna Hora / Madison Berry / A Yousef Abuahmad / Jordan Sprenz / Margaret Deyton / Victoria Stalls / Megan Kopp / Allen L Hsu / Mario J Borgnia / Guillaume B E Stewart-Jones / Matthew S Lee / Naomi Bronkema / M Anthony Moody / Kevin Wiehe / Todd Bradley / S Munir Alam / Robert J Parks / Andrew Foulger / Thomas Oguin / Gregory D Sempowski / Mattia Bonsignori / Celia C LaBranche / David C Montefiori / Michael Seaman / Sampa Santra / John Perfect / Joseph R Francica / Geoffrey M Lynn / Baptiste Aussedat / William E Walkowicz / Richard Laga / Garnett Kelsoe / Kevin O Saunders / Daniela Fera / Peter D Kwong / Robert A Seder / Alberto Bartesaghi / George M Shaw / Priyamvada Acharya / Barton F Haynes /   Abstract: Natural antibodies (Abs) can target host glycans on the surface of pathogens. We studied the evolution of glycan-reactive B cells of rhesus macaques and humans using glycosylated HIV-1 envelope (Env) ...Natural antibodies (Abs) can target host glycans on the surface of pathogens. We studied the evolution of glycan-reactive B cells of rhesus macaques and humans using glycosylated HIV-1 envelope (Env) as a model antigen. 2G12 is a broadly neutralizing Ab (bnAb) that targets a conserved glycan patch on Env of geographically diverse HIV-1 strains using a unique heavy-chain (V) domain-swapped architecture that results in fragment antigen-binding (Fab) dimerization. Here, we describe HIV-1 Env Fab-dimerized glycan (FDG)-reactive bnAbs without V-swapped domains from simian-human immunodeficiency virus (SHIV)-infected macaques. FDG Abs also recognized cell-surface glycans on diverse pathogens, including yeast and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) spike. FDG precursors were expanded by glycan-bearing immunogens in macaques and were abundant in HIV-1-naive humans. Moreover, FDG precursors were predominately mutated IgMIgDCD27, thus suggesting that they originated from a pool of antigen-experienced IgM or marginal zone B cells. #1:  Journal: bioRxiv / Year: 2020 Journal: bioRxiv / Year: 2020Title: Fab-dimerized glycan-reactive antibodies neutralize HIV and are prevalent in humans and rhesus macaques Authors: Williams WB / Meyerhoff RR / Edwards RJ / Li H / Nicely NI / Henderson R / Zhou Y / Janowska K / Mansouri K / Manne K / Stalls V / Hsu AL / Borgnia MJ / Stewart-Jones G / Lee MS / Bronkema N ...Authors: Williams WB / Meyerhoff RR / Edwards RJ / Li H / Nicely NI / Henderson R / Zhou Y / Janowska K / Mansouri K / Manne K / Stalls V / Hsu AL / Borgnia MJ / Stewart-Jones G / Lee MS / Bronkema N / Perfect J / Moody MA / Wiehe K / Bradley T / Kepler TB / Alam SM / Foulger A / Bonsignori M / LaBranche CC / Montefiori DC / Seaman M / Santra S / Francica JR / Lynn GM / Aussedat B / Walkowicz WE / Laga R / Kelso G / Saunders KO / Fera D / Kwong PD / Seder RA / Bartesaghi A / Shaw GM / Acharya P / Haynes BF | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_23145.map.gz emd_23145.map.gz | 5 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-23145-v30.xml emd-23145-v30.xml emd-23145.xml emd-23145.xml | 29.5 KB 29.5 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_23145_fsc.xml emd_23145_fsc.xml | 8.9 KB | Display |  FSC data file FSC data file |
| Images |  emd_23145.png emd_23145.png | 324.4 KB | ||
| Masks |  emd_23145_msk_1.map emd_23145_msk_1.map emd_23145_msk_2.map emd_23145_msk_2.map | 64 MB 64 MB |  Mask map Mask map | |
| Others |  emd_23145_additional_1.map.gz emd_23145_additional_1.map.gz emd_23145_additional_2.map.gz emd_23145_additional_2.map.gz emd_23145_half_map_1.map.gz emd_23145_half_map_1.map.gz emd_23145_half_map_2.map.gz emd_23145_half_map_2.map.gz | 1 MB 31.2 MB 59.4 MB 59.4 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-23145 http://ftp.pdbj.org/pub/emdb/structures/EMD-23145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23145 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-23145 | HTTPS FTP |
-Validation report
| Summary document |  emd_23145_validation.pdf.gz emd_23145_validation.pdf.gz | 531.1 KB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_23145_full_validation.pdf.gz emd_23145_full_validation.pdf.gz | 530.7 KB | Display | |
| Data in XML |  emd_23145_validation.xml.gz emd_23145_validation.xml.gz | 16.2 KB | Display | |
| Data in CIF |  emd_23145_validation.cif.gz emd_23145_validation.cif.gz | 21 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23145 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23145 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23145 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-23145 | HTTPS FTP |
-Related structure data
| Related structure data |  7l6mMC  6vtuC  6xrjC  7l02C  7l06C  7l09C  7l6oC  7lu9C  7luaC C: citing same article ( M: atomic model generated by this map |
|---|---|
| Similar structure data |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|
- Map
Map
| File |  Download / File: emd_23145.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_23145.map.gz / Format: CCP4 / Size: 64 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Final density modified, masked, locally refined map of DH898.1 Fab dimer | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.0665 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
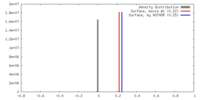
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
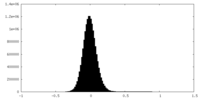
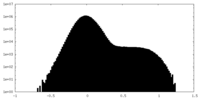
-Mask #1
| File |  emd_23145_msk_1.map emd_23145_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
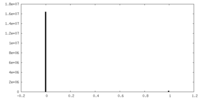
| Projections & Slices |
| ||||||||||||
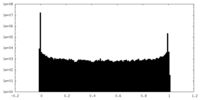
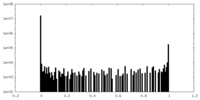
| Density Histograms |
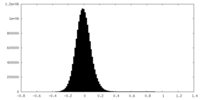
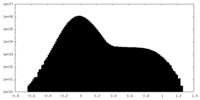
-Mask #2
| File |  emd_23145_msk_2.map emd_23145_msk_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
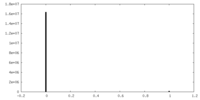
| Projections & Slices |
| ||||||||||||
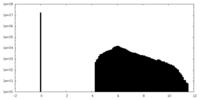
| Density Histograms |
-Additional map: Local resolution of locally refined map of DH898.1 Fab dimer
| File | emd_23145_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Local resolution of locally refined map of DH898.1 Fab dimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
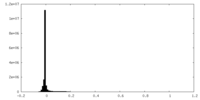
| Density Histograms |
-Additional map: Unmodified locally refined map of DH898.1 Fab dimer
| File | emd_23145_additional_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Unmodified locally refined map of DH898.1 Fab dimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
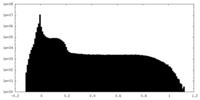
| Density Histograms |
-Half map: Half-map A of locally refined map of DH898.1 Fab dimer
| File | emd_23145_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map A of locally refined map of DH898.1 Fab dimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
-Half map: Half-map B of locally refined map of DH898.1 Fab dimer
| File | emd_23145_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Half-map B of locally refined map of DH898.1 Fab dimer | ||||||||||||
| Projections & Slices |
| ||||||||||||
| Density Histograms |
- Sample components
Sample components
-Entire : Antibody DH898.1 Fab-dimer bound to HIV Env CH848.3.D0949.10.17ch...
| Entire | Name: Antibody DH898.1 Fab-dimer bound to HIV Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 trimer |
|---|---|
| Components |
|
-Supramolecule #1: Antibody DH898.1 Fab-dimer bound to HIV Env CH848.3.D0949.10.17ch...
| Supramolecule | Name: Antibody DH898.1 Fab-dimer bound to HIV Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 trimer type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#3 |
|---|---|
| Molecular weight | Theoretical: 94.2 KDa |
-Supramolecule #2: Antibody DH898.1 Fab-dimer
| Supramolecule | Name: Antibody DH898.1 Fab-dimer / type: complex / ID: 2 / Parent: 1 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  |
| Recombinant expression | Organism:  Homo sapiens (human) / Recombinant cell: 293F Homo sapiens (human) / Recombinant cell: 293F |
-Supramolecule #3: HIV Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 trimer
| Supramolecule | Name: HIV Env CH848.3.D0949.10.17chim.6R.DS.SOSIP.664 trimer type: complex / ID: 3 / Parent: 1 / Macromolecule list: #3 |
|---|---|
| Source (natural) | Organism:   Human immunodeficiency virus 1 Human immunodeficiency virus 1 |
| Recombinant expression | Organism:  Homo sapiens (human) Homo sapiens (human) |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Concentration | 1.5 mg/mL | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Buffer | pH: 7.5 Component:
| |||||||||
| Grid | Model: Quantifoil R1.2/1.3 / Support film - Material: CARBON / Support film - topology: HOLEY ARRAY / Support film - Film thickness: 12.0 nm / Pretreatment - Type: GLOW DISCHARGE | |||||||||
| Vitrification | Cryogen name: ETHANE / Chamber humidity: 95 % / Chamber temperature: 295 K / Instrument: LEICA EM GP Details: 2.5 microliters of sample applied to grid and incubated for 30 seconds in chamber, blotted for 2.5 seconds, and plunge frozen in liquid ethane. | |||||||||
| Details | For the DH898.1-bound complex, purified HIV-1 Env was mixed with 6-fold molar excess of recombinantly expressed and purified DH898.1 Fab to obtain a final Env concentration of 1.5 mg/ml, and incubated for 2 hours at room temperature. |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 (6k x 4k) / Number real images: 3230 / Average exposure time: 4.0 sec. / Average electron dose: 60.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Nominal defocus max: 3.5 µm / Nominal defocus min: 1.0 µm / Nominal magnification: 22500 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
 Movie
Movie Controller
Controller



















 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)