+ Open data
Open data
- Basic information
Basic information
| Entry |  | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | KtrAB complex | |||||||||
 Map data Map data | KtrAB complex of KtrA8 ring with KtrB dimer on each side | |||||||||
 Sample Sample |
| |||||||||
 Keywords Keywords | potassium transporter / membrane transport protein / MEMBRANE PROTEIN | |||||||||
| Function / homology |  Function and homology information Function and homology informationpotassium:chloride symporter activity / monoatomic cation transmembrane transporter activity / potassium ion transport / ATP binding / plasma membrane Similarity search - Function | |||||||||
| Biological species |  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) | |||||||||
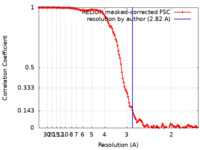
| Method | single particle reconstruction / cryo EM / Resolution: 2.82 Å | |||||||||
 Authors Authors | Vonck J / Stautz J | |||||||||
| Funding support |  Germany, 2 items Germany, 2 items
| |||||||||
 Citation Citation |  Journal: To Be Published Journal: To Be PublishedTitle: KtrAB complex Authors: Stautz J | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Supplemental images |
|---|
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_14851.map.gz emd_14851.map.gz | 161.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-14851-v30.xml emd-14851-v30.xml emd-14851.xml emd-14851.xml | 20.2 KB 20.2 KB | Display Display |  EMDB header EMDB header |
| FSC (resolution estimation) |  emd_14851_fsc.xml emd_14851_fsc.xml | 12.8 KB | Display |  FSC data file FSC data file |
| Images |  emd_14851.png emd_14851.png | 176.9 KB | ||
| Masks |  emd_14851_msk_1.map emd_14851_msk_1.map | 178 MB |  Mask map Mask map | |
| Filedesc metadata |  emd-14851.cif.gz emd-14851.cif.gz | 6.4 KB | ||
| Others |  emd_14851_additional_1.map.gz emd_14851_additional_1.map.gz emd_14851_half_map_1.map.gz emd_14851_half_map_1.map.gz emd_14851_half_map_2.map.gz emd_14851_half_map_2.map.gz | 165.9 MB 139.5 MB 139.5 MB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-14851 http://ftp.pdbj.org/pub/emdb/structures/EMD-14851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14851 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-14851 | HTTPS FTP |
-Validation report
| Summary document |  emd_14851_validation.pdf.gz emd_14851_validation.pdf.gz | 1.2 MB | Display |  EMDB validaton report EMDB validaton report |
|---|---|---|---|---|
| Full document |  emd_14851_full_validation.pdf.gz emd_14851_full_validation.pdf.gz | 1.2 MB | Display | |
| Data in XML |  emd_14851_validation.xml.gz emd_14851_validation.xml.gz | 19.6 KB | Display | |
| Data in CIF |  emd_14851_validation.cif.gz emd_14851_validation.cif.gz | 25.5 KB | Display | |
| Arichive directory |  https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14851 https://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14851 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14851 ftp://ftp.pdbj.org/pub/emdb/validation_reports/EMD-14851 | HTTPS FTP |
-Related structure data
| Related structure data |  7zp9MC M: atomic model generated by this map C: citing same article ( |
|---|---|
| Similar structure data | Similarity search - Function & homology  F&H Search F&H Search |
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_14851.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_14851.map.gz / Format: CCP4 / Size: 178 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | KtrAB complex of KtrA8 ring with KtrB dimer on each side | ||||||||||||||||||||||||||||||||||||
| Projections & slices | Image control
Images are generated by Spider. | ||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 0.831 Å | ||||||||||||||||||||||||||||||||||||
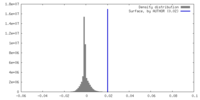
| Density |
| ||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
|
-Supplemental data
-Mask #1
| File |  emd_14851_msk_1.map emd_14851_msk_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
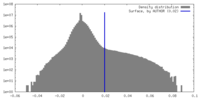
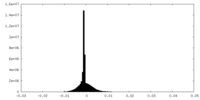
| Density Histograms |
-Additional map: Map at 2.54 Angstrom resolution after density modification in Phenix
| File | emd_14851_additional_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Map at 2.54 Angstrom resolution after density modification in Phenix | ||||||||||||
| Projections & Slices |
| ||||||||||||
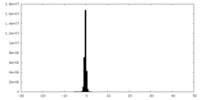
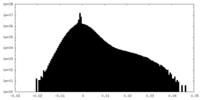
| Density Histograms |
-Half map: #2
| File | emd_14851_half_map_1.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
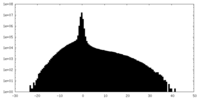
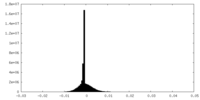
| Density Histograms |
-Half map: #1
| File | emd_14851_half_map_2.map | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Projections & Slices |
| ||||||||||||
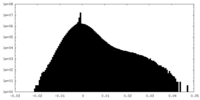
| Density Histograms |
- Sample components
Sample components
-Entire : KtrAB complex with a second KtrB dimer attached
| Entire | Name: KtrAB complex with a second KtrB dimer attached |
|---|---|
| Components |
|
-Supramolecule #1: KtrAB complex with a second KtrB dimer attached
| Supramolecule | Name: KtrAB complex with a second KtrB dimer attached / type: complex / ID: 1 / Parent: 0 / Macromolecule list: #1-#2 |
|---|---|
| Source (natural) | Organism:  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) |
| Molecular weight | Theoretical: 390 KDa |
-Macromolecule #1: Ktr system potassium uptake protein A
| Macromolecule | Name: Ktr system potassium uptake protein A / type: protein_or_peptide / ID: 1 / Number of copies: 8 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) |
| Molecular weight | Theoretical: 23.83692 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MKTGDKQFAV IGLGRFGLAV CKELQDSGSQ VLAVDINEDR VKEAAGFVSQ AIVANCTHEE TVAELKLDDY DMVMIAIGAD VNASILATL IAKEAGVKSV WVKANDRFQA RVLQKIGADH IIMPERDMGI RVARKMLDKR VLEFHPLGSG LAMTEFVVGS R LMGKTLSD ...String: MKTGDKQFAV IGLGRFGLAV CKELQDSGSQ VLAVDINEDR VKEAAGFVSQ AIVANCTHEE TVAELKLDDY DMVMIAIGAD VNASILATL IAKEAGVKSV WVKANDRFQA RVLQKIGADH IIMPERDMGI RVARKMLDKR VLEFHPLGSG LAMTEFVVGS R LMGKTLSD LALCKVEGVQ VLGYKRGPEI IKAPDMSTTL EIGDLIIVVG PQDKLANKLK SL UniProtKB: Ktr system potassium uptake protein A |
-Macromolecule #2: Ktr system potassium uptake protein B
| Macromolecule | Name: Ktr system potassium uptake protein B / type: protein_or_peptide / ID: 2 / Number of copies: 4 / Enantiomer: LEVO |
|---|---|
| Source (natural) | Organism:  Vibrio alginolyticus (bacteria) Vibrio alginolyticus (bacteria) |
| Molecular weight | Theoretical: 49.707715 KDa |
| Recombinant expression | Organism:  |
| Sequence | String: MTQFHQRGVF YVPDGKRDKA KGGEPRIILL SFLGVLLPSA VLLTLPVFSV SGLSITDALF TATSAISVTG LGVVDTGQHF TLAGKILLM CLMQIGGLGQ MTLSAVLLYM FGVRLSLRQQ ALAKEALGQE RQVNLRRLVK KIVTFALVAE AIGFVFLSYR W VPEMGWQT ...String: MTQFHQRGVF YVPDGKRDKA KGGEPRIILL SFLGVLLPSA VLLTLPVFSV SGLSITDALF TATSAISVTG LGVVDTGQHF TLAGKILLM CLMQIGGLGQ MTLSAVLLYM FGVRLSLRQQ ALAKEALGQE RQVNLRRLVK KIVTFALVAE AIGFVFLSYR W VPEMGWQT GMFYALFHSI SAFNNAGFAL FSDSMMSFVN DPLVSFTLAG LFIFGGLGFT VIGDVWRHWR KGFHFLHIHT KI MLIATPL LLLVGTVLFW LLERHNPNTM GSLTTGGQWL AAFFQSASAR TAGFNSVDLT QFTQPALLIM IVLMLIGAGS TST GGGIKV STFAVAFMAT WTFLRQKKHV VMFKRTVNWP TVTKSLAIIV VSGAILTTAM FLLMLTEKAS FDKVMFETIS AFAT VGLTA GLTAELSEPG KYIMIVVMII GRIGPLTLAY MLARPEPTLI KYPEDTVLTG UniProtKB: Ktr system potassium uptake protein B |
-Macromolecule #3: MAGNESIUM ION
| Macromolecule | Name: MAGNESIUM ION / type: ligand / ID: 3 / Number of copies: 8 / Formula: MG |
|---|---|
| Molecular weight | Theoretical: 24.305 Da |
-Macromolecule #4: ADENOSINE-5'-DIPHOSPHATE
| Macromolecule | Name: ADENOSINE-5'-DIPHOSPHATE / type: ligand / ID: 4 / Number of copies: 8 / Formula: ADP |
|---|---|
| Molecular weight | Theoretical: 427.201 Da |
| Chemical component information |  ChemComp-ADP: |
-Macromolecule #5: DODECYL-BETA-D-MALTOSIDE
| Macromolecule | Name: DODECYL-BETA-D-MALTOSIDE / type: ligand / ID: 5 / Number of copies: 36 / Formula: LMT |
|---|---|
| Molecular weight | Theoretical: 510.615 Da |
| Chemical component information |  ChemComp-LMT: |
-Macromolecule #6: POTASSIUM ION
| Macromolecule | Name: POTASSIUM ION / type: ligand / ID: 6 / Number of copies: 4 / Formula: K |
|---|---|
| Molecular weight | Theoretical: 39.098 Da |
-Macromolecule #7: water
| Macromolecule | Name: water / type: ligand / ID: 7 / Number of copies: 43 / Formula: HOH |
|---|---|
| Molecular weight | Theoretical: 18.015 Da |
| Chemical component information |  ChemComp-HOH: |
-Experimental details
-Structure determination
| Method | cryo EM |
|---|---|
 Processing Processing | single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 8 |
|---|---|
| Grid | Model: C-flat-1.2/1.3 / Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: FEI VITROBOT MARK IV |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Image recording | Film or detector model: GATAN K3 BIOQUANTUM (6k x 4k) / Number grids imaged: 1 / Number real images: 2716 / Average electron dose: 40.0 e/Å2 |
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Calibrated defocus max: 2.5 µm / Calibrated defocus min: 1.0 µm / Calibrated magnification: 60168 / Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD / Cs: 2.7 mm / Nominal defocus max: 2.3000000000000003 µm / Nominal defocus min: 1.2 µm / Nominal magnification: 105000 |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER / Cooling holder cryogen: NITROGEN |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
+ Image processing
Image processing
-Atomic model buiding 1
| Details | The model was refined by phenix.real-space-refine |
|---|---|
| Refinement | Space: REAL / Protocol: OTHER |
| Output model |  PDB-7zp9: |
 Movie
Movie Controller
Controller







 Z (Sec.)
Z (Sec.) Y (Row.)
Y (Row.) X (Col.)
X (Col.)