+ Open data
Open data
- Basic information
Basic information
| Entry | Database: EMDB / ID: EMD-3603 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Title | KaiCB circadian clock backbone model based on a Cryo-EM density | |||||||||
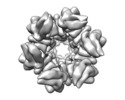
 Map data Map data | Circadian Clock Complex KaiBC | |||||||||
 Sample Sample |
| |||||||||
| Function / homology |  Function and homology information Function and homology informationregulation of phosphorelay signal transduction system / negative regulation of circadian rhythm / entrainment of circadian clock / negative regulation of phosphorylation / protein serine/threonine/tyrosine kinase activity /  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement /  regulation of circadian rhythm / regulation of circadian rhythm /  circadian rhythm / circadian rhythm /  non-specific serine/threonine protein kinase / non-specific serine/threonine protein kinase /  phosphorylation ...regulation of phosphorelay signal transduction system / negative regulation of circadian rhythm / entrainment of circadian clock / negative regulation of phosphorylation / protein serine/threonine/tyrosine kinase activity / phosphorylation ...regulation of phosphorelay signal transduction system / negative regulation of circadian rhythm / entrainment of circadian clock / negative regulation of phosphorylation / protein serine/threonine/tyrosine kinase activity /  Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement / Hydrolases; Acting on acid anhydrides; Acting on acid anhydrides to facilitate cellular and subcellular movement /  regulation of circadian rhythm / regulation of circadian rhythm /  circadian rhythm / circadian rhythm /  non-specific serine/threonine protein kinase / non-specific serine/threonine protein kinase /  phosphorylation / protein serine kinase activity / protein serine/threonine kinase activity / regulation of DNA-templated transcription / magnesium ion binding / phosphorylation / protein serine kinase activity / protein serine/threonine kinase activity / regulation of DNA-templated transcription / magnesium ion binding /  ATP hydrolysis activity / ATP hydrolysis activity /  DNA binding / DNA binding /  ATP binding / identical protein binding / ATP binding / identical protein binding /  plasma membrane / plasma membrane /  cytoplasm cytoplasmSimilarity search - Function | |||||||||
| Biological species |   Synechococcus elongatus (bacteria) Synechococcus elongatus (bacteria) | |||||||||
| Method |  single particle reconstruction / single particle reconstruction /  cryo EM / Resolution: 7.0 Å cryo EM / Resolution: 7.0 Å | |||||||||
 Authors Authors | Schuller JM / Snijder J / Loessl P / Heck AJR / Foerster F | |||||||||
 Citation Citation |  Journal: Science / Year: 2017 Journal: Science / Year: 2017Title: Structures of the cyanobacterial circadian oscillator frozen in a fully assembled state. Authors: Joost Snijder / Jan M Schuller / Anika Wiegard / Philip Lössl / Nicolas Schmelling / Ilka M Axmann / Jürgen M Plitzko / Friedrich Förster / Albert J R Heck /   Abstract: Cyanobacteria have a robust circadian oscillator, known as the Kai system. Reconstituted from the purified protein components KaiC, KaiB, and KaiA, it can tick autonomously in the presence of ...Cyanobacteria have a robust circadian oscillator, known as the Kai system. Reconstituted from the purified protein components KaiC, KaiB, and KaiA, it can tick autonomously in the presence of adenosine 5'-triphosphate (ATP). The KaiC hexamers enter a natural 24-hour reaction cycle of autophosphorylation and assembly with KaiB and KaiA in numerous diverse forms. We describe the preparation of stoichiometrically well-defined assemblies of KaiCB and KaiCBA, as monitored by native mass spectrometry, allowing for a structural characterization by single-particle cryo-electron microscopy and mass spectrometry. Our data reveal details of the interactions between the Kai proteins and provide a structural basis to understand periodic assembly of the protein oscillator. | |||||||||
| History |
|
- Structure visualization
Structure visualization
| Movie |
 Movie viewer Movie viewer |
|---|---|
| Structure viewer | EM map:  SurfView SurfView Molmil Molmil Jmol/JSmol Jmol/JSmol |
| Supplemental images |
- Downloads & links
Downloads & links
-EMDB archive
| Map data |  emd_3603.map.gz emd_3603.map.gz | 2.6 MB |  EMDB map data format EMDB map data format | |
|---|---|---|---|---|
| Header (meta data) |  emd-3603-v30.xml emd-3603-v30.xml emd-3603.xml emd-3603.xml | 10 KB 10 KB | Display Display |  EMDB header EMDB header |
| Images |  emd_3603.png emd_3603.png | 73.2 KB | ||
| Archive directory |  http://ftp.pdbj.org/pub/emdb/structures/EMD-3603 http://ftp.pdbj.org/pub/emdb/structures/EMD-3603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3603 ftp://ftp.pdbj.org/pub/emdb/structures/EMD-3603 | HTTPS FTP |
-Related structure data
- Links
Links
| EMDB pages |  EMDB (EBI/PDBe) / EMDB (EBI/PDBe) /  EMDataResource EMDataResource |
|---|---|
| Related items in Molecule of the Month |
- Map
Map
| File |  Download / File: emd_3603.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) Download / File: emd_3603.map.gz / Format: CCP4 / Size: 28.7 MB / Type: IMAGE STORED AS FLOATING POINT NUMBER (4 BYTES) | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Annotation | Circadian Clock Complex KaiBC | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Voxel size | X=Y=Z: 1.35 Å | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Density |
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Symmetry | Space group: 1 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| Details | EMDB XML:
CCP4 map header:
| ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
-Supplemental data
- Sample components
Sample components
-Entire : KaiCB circadian clock complex
| Entire | Name: KaiCB circadian clock complex |
|---|---|
| Components |
|
-Supramolecule #1: KaiCB circadian clock complex
| Supramolecule | Name: KaiCB circadian clock complex / type: complex / ID: 1 / Parent: 0 |
|---|---|
| Source (natural) | Organism:   Synechococcus elongatus (bacteria) Synechococcus elongatus (bacteria) |
| Recombinant expression | Organism:   Synechococcus elongatus (bacteria) Synechococcus elongatus (bacteria) |
-Experimental details
-Structure determination
| Method |  cryo EM cryo EM |
|---|---|
 Processing Processing |  single particle reconstruction single particle reconstruction |
| Aggregation state | particle |
- Sample preparation
Sample preparation
| Buffer | pH: 7.5 |
|---|---|
| Grid | Material: COPPER / Pretreatment - Type: GLOW DISCHARGE |
| Vitrification | Cryogen name: ETHANE / Instrument: HOMEMADE PLUNGER |
- Electron microscopy
Electron microscopy
| Microscope | FEI TITAN KRIOS |
|---|---|
| Electron beam | Acceleration voltage: 300 kV / Electron source:  FIELD EMISSION GUN FIELD EMISSION GUN |
| Electron optics | Illumination mode: FLOOD BEAM / Imaging mode: BRIGHT FIELD Bright-field microscopy Bright-field microscopy |
| Sample stage | Specimen holder model: FEI TITAN KRIOS AUTOGRID HOLDER |
| Image recording | Film or detector model: GATAN K2 SUMMIT (4k x 4k) / Detector mode: COUNTING / Average exposure time: 15.2 sec. / Average electron dose: 45.0 e/Å2 |
| Experimental equipment |  Model: Titan Krios / Image courtesy: FEI Company |
- Image processing
Image processing
| CTF correction | Software - Name: CTFFIND (ver. 4) |
|---|---|
| Startup model | Type of model: INSILICO MODEL In silico model: 3D Shape with the approximate demensions of the 2D Classes |
| Initial angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 1.4) |
| Final angle assignment | Type: PROJECTION MATCHING / Software - Name: RELION (ver. 1.4) |
| Final reconstruction | Resolution.type: BY AUTHOR / Resolution: 7.0 Å / Resolution method: FSC 0.143 CUT-OFF / Number images used: 5137 |
-Atomic model buiding 1
| Refinement | Protocol: RIGID BODY FIT |
|---|
 Movie
Movie Controller
Controller