+Search query
-Structure paper
| Title | Cryo-EM structures reveal the activation and substrate recognition mechanism of human enteropeptidase. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 13, Issue 1, Page 6955, Year 2022 |
| Publish date | Nov 14, 2022 |
 Authors Authors | Xiaoli Yang / Zhanyu Ding / Lisi Peng / Qiuyue Song / Deyu Zhang / Fang Cui / Chuanchao Xia / Keliang Li / Hua Yin / Shiyu Li / Zhaoshen Li / Haojie Huang /  |
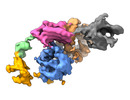
| PubMed Abstract | Enteropeptidase (EP) initiates intestinal digestion by proteolytically processing trypsinogen, generating catalytically active trypsin. EP dysfunction causes a series of pancreatic diseases including ...Enteropeptidase (EP) initiates intestinal digestion by proteolytically processing trypsinogen, generating catalytically active trypsin. EP dysfunction causes a series of pancreatic diseases including acute necrotizing pancreatitis. However, the molecular mechanisms of EP activation and substrate recognition remain elusive, due to the lack of structural information on the EP heavy chain. Here, we report cryo-EM structures of human EP in inactive, active, and substrate-bound states at resolutions from 2.7 to 4.9 Å. The EP heavy chain was observed to clamp the light chain with CUB2 domain for substrate recognition. The EP light chain N-terminus induced a rearrangement of surface-loops from inactive to active conformations, resulting in activated EP. The heavy chain then served as a hinge for light-chain conformational changes to recruit and subsequently cleave substrate. Our study provides structural insights into rearrangements of EP surface-loops and heavy chain dynamics in the EP catalytic cycle, advancing our understanding of EP-associated pancreatitis. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:36376282 / PubMed:36376282 /  PubMed Central PubMed Central |
| Methods | EM (single particle) |
| Resolution | 2.7 - 4.9 Å |
| Structure data | EMDB-32714, PDB-7wqw: EMDB-32715, PDB-7wqx: EMDB-32716, PDB-7wqz: EMDB-32717, PDB-7wr7: EMDB-32828: Inhibited EP-complete EMDB-32829: Substrate bound EP |
| Chemicals |  ChemComp-NAG: 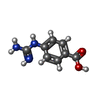 ChemComp-GBS: |
| Source |
|
 Keywords Keywords |  MEMBRANE PROTEIN / MEMBRANE PROTEIN /  COMPLEX / COMPLEX /  HYDROLASE / HYDROLASE /  MEMBRANE PROTEIN/HYDROLASE / MEMBRANE PROTEIN/HYDROLASE /  MEMBRANE PROTEIN-HYDROLASE complex MEMBRANE PROTEIN-HYDROLASE complex |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN Papers
About EMN Papers