+Search query
-Structure paper
| Title | Structural and functional analysis of the promiscuous AcrB and AdeB efflux pumps suggests different drug binding mechanisms. |
|---|---|
| Journal, issue, pages | Nat Commun, Vol. 12, Issue 1, Page 6919, Year 2021 |
| Publish date | Nov 25, 2021 |
 Authors Authors | Alina Ornik-Cha / Julia Wilhelm / Jessica Kobylka / Hanno Sjuts / Attilio V Vargiu / Giuliano Malloci / Julian Reitz / Anja Seybert / Achilleas S Frangakis / Klaas M Pos /   |
| PubMed Abstract | Upon antibiotic stress Gram-negative pathogens deploy resistance-nodulation-cell division-type tripartite efflux pumps. These include a H/drug antiporter module that recognizes structurally diverse ...Upon antibiotic stress Gram-negative pathogens deploy resistance-nodulation-cell division-type tripartite efflux pumps. These include a H/drug antiporter module that recognizes structurally diverse substances, including antibiotics. Here, we show the 3.5 Å structure of subunit AdeB from the Acinetobacter baumannii AdeABC efflux pump solved by single-particle cryo-electron microscopy. The AdeB trimer adopts mainly a resting state with all protomers in a conformation devoid of transport channels or antibiotic binding sites. However, 10% of the protomers adopt a state where three transport channels lead to the closed substrate (deep) binding pocket. A comparison between drug binding of AdeB and Escherichia coli AcrB is made via activity analysis of 20 AdeB variants, selected on basis of side chain interactions with antibiotics observed in the AcrB periplasmic domain X-ray co-structures with fusidic acid (2.3 Å), doxycycline (2.1 Å) and levofloxacin (2.7 Å). AdeABC, compared to AcrAB-TolC, confers higher resistance to E. coli towards polyaromatic compounds and lower resistance towards antibiotic compounds. |
 External links External links |  Nat Commun / Nat Commun /  PubMed:34824229 / PubMed:34824229 /  PubMed Central PubMed Central |
| Methods | EM (single particle) / X-ray diffraction |
| Resolution | 2.1 - 3.84 Å |
| Structure data | EMDB-12088, PDB-7b8p: EMDB-12089, PDB-7b8q:  PDB-7b8r:  PDB-7b8s:  PDB-7b8t: |
| Chemicals |  ChemComp-PEG:  ChemComp-DXT:  ChemComp-HOH: 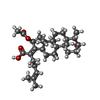 ChemComp-FUA: 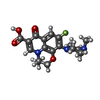 ChemComp-LFX: |
| Source |
|
 Keywords Keywords |  TRANSPORT PROTEIN / RND-transporter / multidrug transporter / TRANSPORT PROTEIN / RND-transporter / multidrug transporter /  antibiotic resistance / antibiotic resistance /  membrane protein / RND transporter / membrane protein / RND transporter /  multidrug resistance / multidrug resistance /  efflux pump / efflux pump /  antibiotics antibiotics |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN Papers
About EMN Papers









