+Search query
-Structure paper
| Title | Structures of the AMPA receptor in complex with its auxiliary subunit cornichon. |
|---|---|
| Journal, issue, pages | Science, Vol. 366, Issue 6470, Page 1259-1263, Year 2019 |
| Publish date | Dec 6, 2019 |
 Authors Authors | Terunaga Nakagawa /  |
| PubMed Abstract | In the brain, AMPA-type glutamate receptors (AMPARs) form complexes with their auxiliary subunits and mediate the majority of fast excitatory neurotransmission. Signals transduced by these complexes ...In the brain, AMPA-type glutamate receptors (AMPARs) form complexes with their auxiliary subunits and mediate the majority of fast excitatory neurotransmission. Signals transduced by these complexes are critical for synaptic plasticity, learning, and memory. The two major categories of AMPAR auxiliary subunits are transmembrane AMPAR regulatory proteins (TARPs) and cornichon homologs (CNIHs); these subunits share little homology and play distinct roles in controlling ion channel gating and trafficking of AMPAR. Here, I report high-resolution cryo-electron microscopy structures of AMPAR in complex with CNIH3. Contrary to its predicted membrane topology, CNIH3 lacks an extracellular domain and instead contains four membrane-spanning helices. The protein-protein interaction interface that dictates channel modulation and the lipids surrounding the complex are revealed. These structures provide insights into the molecular mechanism for ion channel modulation and assembly of AMPAR/CNIH3 complexes. |
 External links External links |  Science / Science /  PubMed:31806817 PubMed:31806817 |
| Methods | EM (single particle) |
| Resolution | 2.97 - 4.4 Å |
| Structure data | EMDB-20330, PDB-6peq:  EMDB-20332: EMDB-20654, PDB-6u5s: EMDB-20666, PDB-6u6i:  EMDB-20717: EMDB-20727, PDB-6ucb:  EMDB-20732: EMDB-20733, PDB-6ud4: EMDB-20734, PDB-6ud8: |
| Chemicals | 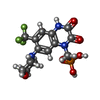 ChemComp-ZK1:  ChemComp-OLC:  ChemComp-PAM:  ChemComp-CLR:  ChemComp-NAG: |
| Source |
|
 Keywords Keywords |  TRANSPORT PROTEIN / TRANSPORT PROTEIN /  ionotropic glutamate receptor / ionotropic glutamate receptor /  AMPA receptor / AMPA receptor /  cornichon / auxiliary subunit / cornichon / auxiliary subunit /  ion channel / ion channel /  ligand gated ion channel / ligand gated ion channel /  synaptic transmission / excitatory synaptic transmission / synaptic transmission / excitatory synaptic transmission /  neurotransmitter receptor / neurotransmitter receptor /  stargazin / TARP / ZK200775 / stargazin / TARP / ZK200775 /  lipid / lipid /  MPQX MPQX |
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN Papers
About EMN Papers

















